Noncontact Atomic Force Microscopy - Yale School of Engineering ...
Noncontact Atomic Force Microscopy - Yale School of Engineering ...
Noncontact Atomic Force Microscopy - Yale School of Engineering ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Tu-0940<br />
Anisotropic Hydration <strong>of</strong> Biological Molecules Visualized by Three-<br />
Dimensional Scanning <strong>Force</strong> <strong>Microscopy</strong><br />
Hitoshi Asakawa 1 , Yasumasa Ueda 1 and Takeshi Fukuma 1,2<br />
1 Frontier Science Organization, Kanazawa University, Japan<br />
2 PRESTO, JST, Japan<br />
Hydration <strong>of</strong> biological molecules has grave impact on important biological processes<br />
such as membrane fusions and protein activities. However, molecular-scale details <strong>of</strong><br />
local hydration structures have remained to be understood due to the lack <strong>of</strong> a method<br />
able to visualize their three-dimensional (3D) distribution at sub-nanometer resolution. In<br />
order to resolve this issue, we have recently developed a novel technique referred to as<br />
3D scanning force microscopy (3D-SFM). Combined with frequency modulation atomic<br />
force microscopy (FM-AFM) in liquid, the method allows us to record 3D volume data <strong>of</strong><br />
frequency shift (Δf ) induced by tip-sample interaction force in less than 1 min. We are<br />
able to slice the obtained 3D-SFM image in any directions to produce 2D cross-sectional<br />
Δf images.<br />
In this study, we have investigated hydration structures formed at the interface between<br />
a model biological membrane and aqueous solution. A dipalmitoylphosphatidylcholine<br />
(DPPC) bilayer was formed on a cleaved mica surface as a model biological membrane in<br />
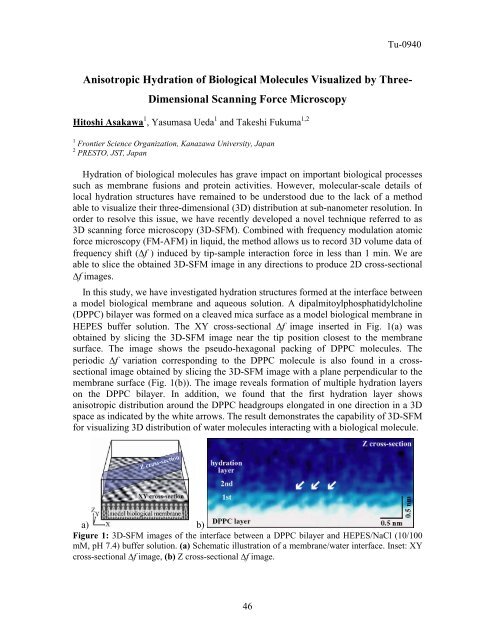
HEPES buffer solution. The XY cross-sectional Δf image inserted in Fig. 1(a) was<br />
obtained by slicing the 3D-SFM image near the tip position closest to the membrane<br />
surface. The image shows the pseudo-hexagonal packing <strong>of</strong> DPPC molecules. The<br />
periodic Δf variation corresponding to the DPPC molecule is also found in a crosssectional<br />
image obtained by slicing the 3D-SFM image with a plane perpendicular to the<br />
membrane surface (Fig. 1(b)). The image reveals formation <strong>of</strong> multiple hydration layers<br />
on the DPPC bilayer. In addition, we found that the first hydration layer shows<br />
anisotropic distribution around the DPPC headgroups elongated in one direction in a 3D<br />
space as indicated by the white arrows. The result demonstrates the capability <strong>of</strong> 3D-SFM<br />
for visualizing 3D distribution <strong>of</strong> water molecules interacting with a biological molecule.<br />
a) b)<br />
Figure 1: 3D-SFM images <strong>of</strong> the interface between a DPPC bilayer and HEPES/NaCl (10/100<br />
mM, pH 7.4) buffer solution. (a) Schematic illustration <strong>of</strong> a membrane/water interface. Inset: XY<br />
cross-sectional Δf image, (b) Z cross-sectional Δf image.<br />
46