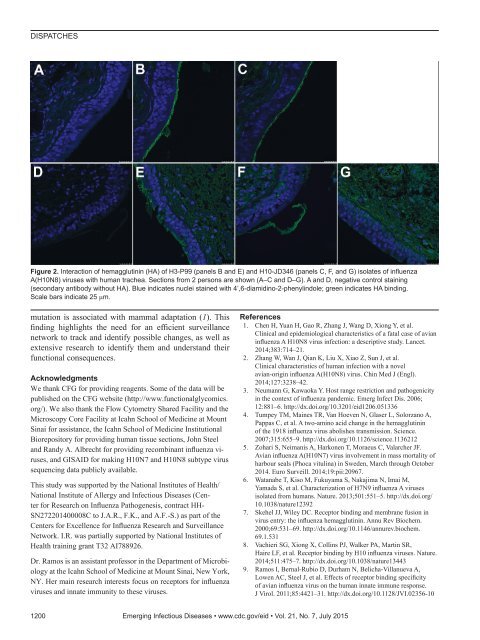
DISPATCHESFigure 2. Interaction of hemagglutinin (HA) of H3-P99 (panels B and E) and H10-JD346 (panels C, F, and G) isolates of influenzaA(H10N8) viruses with human trachea. Sections from 2 persons are shown (A–C and D–G). A and D, negative control staining(secondary antibody without HA). Blue indicates nuclei stained with 4’,6-diamidino-2-phenylindole; green indicates HA binding.Scale bars indicate 25 µm.mutation is associated with mammal adaptation (1). Thisfinding highlights the need for an efficient surveillancenetwork to track and identify possible changes, as well asextensive research to identify them and understand theirfunctional consequences.AcknowledgmentsWe thank CFG for providing reagents. Some of the data will bepublished on the CFG website (http://www.functionalglycomics.org/). We also thank the Flow Cytometry Shared Facility and theMicroscopy Core Facility at Icahn School of Medicine at MountSinai for assistance, the Icahn School of Medicine InstitutionalBiorepository for providing human tissue sections, John Steeland Randy A. Albrecht for providing recombinant influenza viruses,and GISAID for making H10N7 and H10N8 subtype virussequencing data publicly available.This study was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (Centerfor Research on Influenza Pathogenesis, contract HH-SN272201400008C to J.A.R., F.K., and A.F.-S.) as part of theCenters for Excellence for Influenza Research and SurveillanceNetwork. I.R. was partially supported by National Institutes ofHealth training grant T32 AI788926.Dr. Ramos is an assistant professor in the Department of Microbiologyat the Icahn School of Medicine at Mount Sinai, New York,NY. Her main research interests focus on receptors for influenzaviruses and innate immunity to these viruses.References1. Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, et al.Clinical and epidemiological characteristics of a fatal case of avianinfluenza A H10N8 virus infection: a descriptive study. Lancet.2014;383:714–21.2. Zhang W, Wan J, Qian K, Liu X, Xiao Z, Sun J, et al.Clinical characteristics of human infection with a novelavian-origin influenza A(H10N8) virus. Chin Med J (Engl).2014;127:3238–42.3. Neumann G, Kawaoka Y. Host range restriction and pathogenicityin the context of influenza pandemic. Emerg Infect Dis. 2006;12:881–6. http://dx.doi.org/10.3201/eid1206.0513364. Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A,Pappas C, et al. A two-amino acid change in the hemagglutininof the 1918 influenza virus abolishes transmission. Science.2007;315:655–9. http://dx.doi.org/10.1126/science.11362125. Zohari S, Neimanis A, Harkonen T, Moraeus C, Valarcher JF.Avian influenza A(H10N7) virus involvement in mass mortality ofharbour seals (Phoca vitulina) in Sweden, March through October2014. Euro Surveill. 2014;19:pii:20967.6. Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M,Yamada S, et al. Characterization of H7N9 influenza A virusesisolated from humans. Nature. 2013;501:551–5. http://dx.doi.org/10.1038/nature123927. Skehel JJ, Wiley DC. Receptor binding and membrane fusion invirus entry: the influenza hemagglutinin. Annu Rev Biochem.2000;69:531–69. http://dx.doi.org/10.1146/annurev.biochem.69.1.5318. Vachieri SG, Xiong X, Collins PJ, Walker PA, Martin SR,Haire LF, et al. Receptor binding by H10 influenza viruses. Nature.2014;511:475–7. http://dx.doi.org/10.1038/nature134439. Ramos I, Bernal-Rubio D, Durham N, Belicha-Villanueva A,Lowen AC, Steel J, et al. Effects of receptor binding specificityof avian influenza virus on the human innate immune response.J Virol. 2011;85:4421–31. http://dx.doi.org/10.1128/JVI.02356-101200 Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015
Receptor Binding of Influenza A(H10N8) Virus10. Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, etal. H7N9 influenza viruses interact preferentially with alpha2,3-linkedsialic acids and bind weakly to alpha2,6-linked sialic acids. J GenVirol. 2013 94:2417–23. http://dx.doi.org/ 10.1099/vir.0.056184-011. Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P.A carboxy-terminal trimerization domain stabilizes conformationalepitopes on the stalk domain of soluble recombinant hemagglutininsubstrates. PLoS ONE. 2012;7:e43603. http://dx.doi.org/10.1371/journal.pone.004360312. Tharakaraman K, Jayaraman A, Raman R, Viswanathan K,Stebbins NW, Johnson D, et al. Glycan receptor binding of theinfluenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486–93.http://dx.doi.org/10.1016/j.cell.2013.05.03413. Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J.Structure and receptor binding preferences of recombinanthemagglutinins from avian and human H6 and H10 influenza Avirus subtypes. J Virol. 2015 Feb 11;pii: JVI.03456-14.http://dx.doi.org/10.1016/j.virol.2014.12.02414. Wang M, Zhang W, Qi J, Wang F, Zhou J, Bi Y, et al. Structuralbasis for preferential avian receptor binding by the human-infectingH10N8 avian influenza virus. Nat Commun. 2015;6:5600.http://dx.doi.org/10.1038/ncomms660015. Beare AS, Webster RG. Replication of avian influenza viruses inhumans. Arch Virol. 1991;119:37–42. http://dx.doi.org/10.1007/BF01314321Address for correspondence: Irene Ramos, Department ofMicrobiology, Icahn School of Medicine at Mount Sinai, 1468Madison Ave, Box 1124, New York, NY 10029, USA;email: irene.ramos-lopez@mssm.edu; or Florian Krammer,Department of Microbiology, Icahn School of Medicine at MountSinai, 1 Gustave L. Levy Pl, Box 1124, New York, NY 10029, USA;email: florian.krammer@mssm.eduMarch 2015: TuberculosisIncluding:• Evaluation of the Benefits and Risks ofIntroducing Ebola Community Care Centers,Sierra Leone• Nanomicroarray and MultiplexNext-Generation Sequencing forSimultaneous Identification andCharacterization of Influenza Viruses• Multidrug-Resistant Tuberculosis in Europe,2010–2011• Risk Factors for Death from InvasivePneumococcal Disease, Europe, 2010• Mycoplasma pneumoniae and Chlamydiaspp. Infection in Community-AcquiredPneumonia, Germany, 2011–2012• Epidemiology of Human Mycobacteriumbovis Disease, California, USA, 2003–2011• Regional Spread of Ebola Virus, West Africa,2014• Spillover of Mycobacterium bovis fromWildlife to Livestock, South Africa• Prisons as Reservoir for CommunityTransmission of Tuberculosis, Brazil• Polycystic Echinococcosis in Pacas, AmazonRegion, Peru• Red Deer as Maintenance Host for BovineTuberculosis, Alpine Regionhttp://wwwnc.cdc.gov/eid/articles/issue/21/3/table-of-contentsEmerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015 1201
- Page 3 and 4:
July 2015SynopsisOn the CoverMarian
- Page 5 and 6:
1240 Gastroenteritis OutbreaksCause
- Page 7 and 8:
SYNOPSISDisseminated Infections wit
- Page 9 and 10:
Disseminated Infections with Talaro
- Page 11 and 12:
Disseminated Infections with Talaro
- Page 13 and 14:
Macacine Herpesvirus 1 inLong-Taile
- Page 15 and 16:
Macacine Herpesvirus 1 in Macaques,
- Page 17 and 18:
Macacine Herpesvirus 1 in Macaques,
- Page 19:
Macacine Herpesvirus 1 in Macaques,
- Page 23:
Malaria among Young Infants, Africa
- Page 26 and 27:
RESEARCHFigure 3. Dynamics of 19-kD
- Page 28 and 29:
Transdermal Diagnosis of MalariaUsi
- Page 30 and 31:
RESEARCHFigure 2. A) Acoustic trace
- Page 32 and 33:
RESEARCHof malaria-infected mosquit
- Page 34 and 35:
Lack of Transmission amongClose Con
- Page 36 and 37:
RESEARCH(IFA) and microneutralizati
- Page 38 and 39:
RESEARCHoropharyngeal, and serum sa
- Page 40 and 41:
RESEARCH6. Assiri A, McGeer A, Perl
- Page 42 and 43:
RESEARCHadvanced genomic sequencing
- Page 44 and 45:
RESEARCHTable 2. Next-generation se
- Page 46 and 47:
RESEARCHTable 3. Mutation analysis
- Page 48 and 49:
RESEARCHReferences1. Baize S, Panne
- Page 50 and 51:
Parechovirus Genotype 3 Outbreakamo
- Page 52 and 53:
RESEARCHFigure 1. Venn diagramshowi
- Page 54 and 55:
RESEARCHTable 2. HPeV testing of sp
- Page 56 and 57: RESEARCHFigure 5. Distribution of h
- Page 58 and 59: RESEARCHReferences1. Selvarangan R,
- Page 60 and 61: RESEARCHthe left lobe was sampled b
- Page 62 and 63: RESEARCHTable 2. Middle East respir
- Page 64 and 65: RESEARCHseroprevalence in domestic
- Page 66 and 67: RESEARCHmeasure their current surve
- Page 68 and 69: RESEARCHTable 2. States with labora
- Page 70 and 71: RESEARCHFigure 2. Comparison of sur
- Page 72 and 73: RESEARCH9. Centers for Disease Cont
- Page 74 and 75: RESEARCHthe analyses. Cases in pers
- Page 76 and 77: RESEARCHTable 3. Sampling results (
- Page 78 and 79: RESEARCHpresence of Legionella spp.
- Page 80 and 81: Seroprevalence for Hepatitis Eand O
- Page 82 and 83: RESEARCHTable 1. Description of stu
- Page 84 and 85: RESEARCHTable 3. Crude and adjusted
- Page 86 and 87: RESEARCHrates by gender or HIV stat
- Page 88 and 89: RESEARCH25. Taha TE, Kumwenda N, Ka
- Page 90 and 91: POLICY REVIEWDutch Consensus Guidel
- Page 92 and 93: POLICY REVIEWTable 3. Comparison of
- Page 94 and 95: POLICY REVIEW6. Botelho-Nevers E, F
- Page 96 and 97: DISPATCHESFigure 1. Phylogenetic tr
- Page 98 and 99: DISPATCHESSevere Pediatric Adenovir
- Page 100 and 101: DISPATCHESTable 1. Demographics and
- Page 102 and 103: DISPATCHES13. Kim YJ, Hong JY, Lee
- Page 104 and 105: DISPATCHESTable. Alignment of resid
- Page 108 and 109: DISPATCHESSchmallenberg Virus Recur
- Page 110 and 111: DISPATCHESFigure 2. Detection of Sc
- Page 112 and 113: DISPATCHESFigure 1. Histopathologic
- Page 114: DISPATCHESFigure 2. Detection of fo
- Page 117 and 118: Influenza Virus Strains in the Amer
- Page 119 and 120: Novel Arenavirus Isolates from Nama
- Page 121 and 122: Novel Arenaviruses, Southern Africa
- Page 123 and 124: Readability of Ebola Informationon
- Page 125 and 126: Readability of Ebola Information on
- Page 127 and 128: Patients under investigation for ME
- Page 129 and 130: Patients under investigation for ME
- Page 131 and 132: Wildlife Reservoir for Hepatitis E
- Page 133 and 134: Asymptomatic Malaria and Other Infe
- Page 135 and 136: Asymptomatic Malaria in Children fr
- Page 137 and 138: Bufavirus in Wild Shrews and Nonhum
- Page 139 and 140: Bufavirus in Wild Shrews and Nonhum
- Page 141 and 142: Range Expansion for Rat Lungworm in
- Page 143 and 144: Slow Clearance of Plasmodium falcip
- Page 145 and 146: Slow Clearance of Plasmodium falcip
- Page 147 and 148: Gastroenteritis Caused by Norovirus
- Page 149 and 150: Ebola Virus Stability on Surfaces a
- Page 151 and 152: Ebola Virus Stability on Surfaces a
- Page 153 and 154: Outbreak of Ciprofloxacin-Resistant
- Page 155 and 156: Outbreak of S. sonnei, South KoreaT
- Page 157 and 158:
Rapidly Expanding Range of Highly P
- Page 159 and 160:
Cluster of Ebola Virus Disease, Bon
- Page 161 and 162:
Cluster of Ebola Virus Disease, Lib
- Page 163 and 164:
ANOTHER DIMENSIONThe Past Is Never
- Page 165 and 166:
Measles Epidemic, Boston, Massachus
- Page 167 and 168:
LETTERSInfluenza A(H5N6)Virus Reass
- Page 169 and 170:
LETTERSsystem (8 kb-span paired-end
- Page 171 and 172:
LETTERS3. Van Hong N, Amambua-Ngwa
- Page 173 and 174:
LETTERSTable. Prevalence of Bartone
- Page 175 and 176:
LETTERSavian influenza A(H5N1) viru
- Page 177 and 178:
LETTERSprovinces and a total of 200
- Page 179 and 180:
LETTERS7. Manian FA. Bloodstream in
- Page 181 and 182:
LETTERSforward projections. N Engl
- Page 183 and 184:
LETTERS3. Guindon S, Gascuel OA. Si
- Page 185 and 186:
BOOKS AND MEDIAin the port cities o
- Page 187 and 188:
ABOUT THE COVERNorth was not intere
- Page 189 and 190:
Earning CME CreditTo obtain credit,
- Page 191:
Emerging Infectious Diseases is a p