DISPATCHESDistinct Lineages of Bufavirus inWild Shrews and Nonhuman PrimatesMichihito Sasaki, Yasuko Orba,Paulina D. Anindita, Akihiro Ishii, Keisuke Ueno, 1Bernard M. Hang’ombe, Aaron S. Mweene,Kimihito Ito, Hirofumi SawaViral metagenomic analysis identified a new parvovirus genomein the intestinal contents of wild shrews in Zambia.Related viruses were detected in spleen tissues from wildshrews and nonhuman primates. Phylogenetic analysesshowed that these viruses are related to human bufaviruses,highlighting the presence and genetic diversity of bufavirusesin wildlife.Bufavirus (BuV), a recently described parvovirus, wasinitially discovered in the feces of a child with diarrheain Burkina Faso in 2012 (1). Thereafter, BuV was identifiedin fecal samples from children and adults with gastroenteritisin Bhutan, Finland, and the Netherlands in 2014 (2–4),respectively. Genome sequences and phylogenetic analysesrevealed that BuV comprised at least 3 genotypes andwas distinct from all other known members of the Parvoviridaefamily (1,2). The International Committee on Taxonomyof Viruses assigned BuV as a new species of thegenus Protoparvovirus in the subfamily Parvovirinae (5).Whether BuV is an etiologic agent of human gastroenteritisremains unclear, but knowledge about its distribution andgenetic divergence in humans is accumulating. However,whether BuV infection exists in wildlife remains unanswered.Through use of metagenomics, we previously describedthe enteric virome of wild shrews of the Crociduragenus sampled at Mpulungu, Zambia, in 2012 (6). Fromthis sequence dataset (GenBank/EMBL/DDBJ accessionno. DRA002561), we identified sequence reads related toBuV. Here, we describe the genome of this new parvovirus.The StudyWe determined the nearly complete genome sequence ofBuV, which we named Mpulungu BuV (MpBuV), by fillinggenome gaps with primer walking and rapid amplificationof cDNA ends (GenBank/EMBL/DDBJ accession no.AB937988). The MpBuV genome comprises 4,613 nt andAuthor affiliations: Hokkaido University, Sapporo, Japan(M. Sasaki, Y. Orba, P.D. Anindita, A. Ishii, K. Ueno, K. Ito,H. Sawa); University of Zambia, Lusaka, Zambia(B.M. Hang’ombe, A.S. Mweene)DOI: http://dx.doi.org/10.3201/eid2107.141969encodes open reading frames of the nonstructural protein(NS) 1 and the viral capsid protein (VP) 1 and VP2 (onlineTechnical Appendix Figure, http://wwwnc.cdc.gov/EID/article/21/7/14-1969-Techapp1.<strong>pdf</strong>). blastp (http://blast.ncbi.nlm.nih.gov) searches showed that the MpBuV NS1,VP1, and VP2 proteins were closely related to those of humanBuVs and the WUHARV parvovirus (E-value = 0.0).WUHARV parvovirus, identified in rhesus monkeys experimentallyinfected with simian immunodeficiency virusunder laboratory conditions in the United States, was foundto be a closely related to human BuVs (7).In MpBuV, NS1 shares 52.5% aa identity with NS1of human BuV (GenBank accession no. JX027296). Wefound that the amino acid sequence identity of VP1 betweenMpBuV and human BuV is 52.3%, whereas that ofVP2 is 51.4%. Similar to human BuV, MpBuV showedpotential splicing signals in the VP1 coding region. Wealso identified the parvovirus-conserved amino acid motifsin NS1, VP1, and VP2 of MpBuV (online TechnicalAppendix Figure) (8–12). Phylogenetic analysis was performedas described previously (6). A Bayesian phylogenetictree was generated on the basis of the full-lengthNS1 proteins of MpBuV, human BuVs, and representativeparvoviruses. MpBuV clustered with human BuVsand WUHARV parvovirus (online Technical AppendixFigure). According to the species demarcation criteria ofthe International Committee on Taxonomy of Viruses,each parvovirus species encodes an NS1 protein sharing
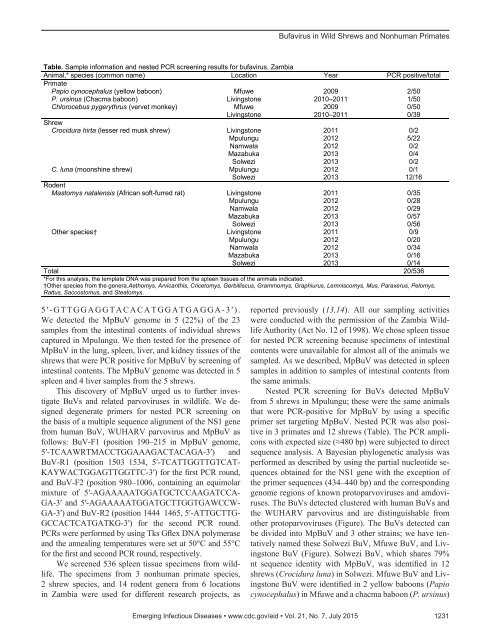
Bufavirus in Wild Shrews and Nonhuman PrimatesTable. Sample information and nested PCR screening results for bufavirus, ZambiaAnimal,* species (common name) Location Year PCR positive/totalPrimatePapio cynocephalus (yellow baboon) Mfuwe 2009 2/50P. ursinus (Chacma baboon) Livingstone 2010–2011 1/50Chlorocebus pygerythrus (vervet monkey) Mfuwe 2009 0/50Livingstone 2010–2011 0/39ShrewCrocidura hirta (lesser red musk shrew) Livingstone 2011 0/2Mpulungu 2012 5/22Namwala 2012 0/2Mazabuka 2013 0/4Solwezi 2013 0/2C. luna (moonshine shrew) Mpulungu 2012 0/1Solwezi 2013 12/16RodentMastomys natalensis (African soft-furred rat) Livingstone 2011 0/35Mpulungu 2012 0/28Namwala 2012 0/29Mazabuka 2013 0/57Solwezi 2013 0/56Other species† Livingstone 2011 0/9Mpulungu 2012 0/20Namwala 2012 0/34Mazabuka 2013 0/16Solwezi 2013 0/14Total 20/536*For this analysis, the template DNA was prepared from the spleen tissues of the animals indicated.†Other species from the genera.Aethomys, Arvicanthis, Cricetomys, Gerbilliscus, Grammomys, Graphiurus, Lemniscomys, Mus, Paraxerus, Pelomys,Rattus, Saccostomus, and Steatomys.5′-GTTGGAGGTACACATGGATGAGGA-3′).We detected the MpBuV genome in 5 (22%) of the 23samples from the intestinal contents of individual shrewscaptured in Mpulungu. We then tested for the presence ofMpBuV in the lung, spleen, liver, and kidney tissues of theshrews that were PCR positive for MpBuV by screening ofintestinal contents. The MpBuV genome was detected in 5spleen and 4 liver samples from the 5 shrews.This discovery of MpBuV urged us to further investigateBuVs and related parvoviruses in wildlife. We designeddegenerate primers for nested PCR screening onthe basis of a multiple sequence alignment of the NS1 genefrom human BuV, WUHARV parvovirus and MpBuV asfollows: BuV-F1 (position 190–215 in MpBuV genome,5′-TCAAWRTMACCTGGAAAGACTACAGA-3′) andBuV-R1 (position 1503 1534, 5′-TCATTGGTTGTCAT-KAYWACTGGAGTTGGTTC-3′) for the first PCR round,and BuV-F2 (position 980–1006, containing an equimolarmixture of 5′-AGAAAAATGGATGCTCCAAGATCCA-GA-3′ and 5′-AGAAAAATGGATGCTTGGTGAWCCW-GA-3′) and BuV-R2 (position 1444 1465, 5′-ATTGCTTG-GCCACTCATGATKG-3′) for the second PCR round.PCRs were performed by using Tks Gflex DNA polymeraseand the annealing temperatures were set at 50°C and 55°Cfor the first and second PCR round, respectively.We screened 536 spleen tissue specimens from wildlife.The specimens from 3 nonhuman primate species,2 shrew species, and 14 rodent genera from 6 locationsin Zambia were used for different research projects, asreported previously (13,14). All our sampling activitieswere conducted with the permission of the Zambia WildlifeAuthority (Act No. 12 of 1998). We chose spleen tissuefor nested PCR screening because specimens of intestinalcontents were unavailable for almost all of the animals wesampled. As we described, MpBuV was detected in spleensamples in addition to samples of intestinal contents fromthe same animals.Nested PCR screening for BuVs detected MpBuVfrom 5 shrews in Mpulungu; these were the same animalsthat were PCR-positive for MpBuV by using a specificprimer set targeting MpBuV. Nested PCR was also positivein 3 primates and 12 shrews (Table). The PCR ampliconswith expected size (≈480 bp) were subjected to directsequence analysis. A Bayesian phylogenetic analysis wasperformed as described by using the partial nucleotide sequencesobtained for the NS1 gene with the exception ofthe primer sequences (434–440 bp) and the correspondinggenome regions of known protoparvoviruses and amdoviruses.The BuVs detected clustered with human BuVs andthe WUHARV parvovirus and are distinguishable fromother protoparvoviruses (Figure). The BuVs detected canbe divided into MpBuV and 3 other strains; we have tentativelynamed these Solwezi BuV, Mfuwe BuV, and LivingstoneBuV (Figure). Solwezi BuV, which shares 79%nt sequence identity with MpBuV, was identified in 12shrews (Crocidura luna) in Solwezi. Mfuwe BuV and LivingstoneBuV were identified in 2 yellow baboons (Papiocynocephalus) in Mfuwe and a chacma baboon (P. ursinus)Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015 1231
- Page 3 and 4:
July 2015SynopsisOn the CoverMarian
- Page 5 and 6:
1240 Gastroenteritis OutbreaksCause
- Page 7 and 8:
SYNOPSISDisseminated Infections wit
- Page 9 and 10:
Disseminated Infections with Talaro
- Page 11 and 12:
Disseminated Infections with Talaro
- Page 13 and 14:
Macacine Herpesvirus 1 inLong-Taile
- Page 15 and 16:
Macacine Herpesvirus 1 in Macaques,
- Page 17 and 18:
Macacine Herpesvirus 1 in Macaques,
- Page 19:
Macacine Herpesvirus 1 in Macaques,
- Page 23:
Malaria among Young Infants, Africa
- Page 26 and 27:
RESEARCHFigure 3. Dynamics of 19-kD
- Page 28 and 29:
Transdermal Diagnosis of MalariaUsi
- Page 30 and 31:
RESEARCHFigure 2. A) Acoustic trace
- Page 32 and 33:
RESEARCHof malaria-infected mosquit
- Page 34 and 35:
Lack of Transmission amongClose Con
- Page 36 and 37:
RESEARCH(IFA) and microneutralizati
- Page 38 and 39:
RESEARCHoropharyngeal, and serum sa
- Page 40 and 41:
RESEARCH6. Assiri A, McGeer A, Perl
- Page 42 and 43:
RESEARCHadvanced genomic sequencing
- Page 44 and 45:
RESEARCHTable 2. Next-generation se
- Page 46 and 47:
RESEARCHTable 3. Mutation analysis
- Page 48 and 49:
RESEARCHReferences1. Baize S, Panne
- Page 50 and 51:
Parechovirus Genotype 3 Outbreakamo
- Page 52 and 53:
RESEARCHFigure 1. Venn diagramshowi
- Page 54 and 55:
RESEARCHTable 2. HPeV testing of sp
- Page 56 and 57:
RESEARCHFigure 5. Distribution of h
- Page 58 and 59:
RESEARCHReferences1. Selvarangan R,
- Page 60 and 61:
RESEARCHthe left lobe was sampled b
- Page 62 and 63:
RESEARCHTable 2. Middle East respir
- Page 64 and 65:
RESEARCHseroprevalence in domestic
- Page 66 and 67:
RESEARCHmeasure their current surve
- Page 68 and 69:
RESEARCHTable 2. States with labora
- Page 70 and 71:
RESEARCHFigure 2. Comparison of sur
- Page 72 and 73:
RESEARCH9. Centers for Disease Cont
- Page 74 and 75:
RESEARCHthe analyses. Cases in pers
- Page 76 and 77:
RESEARCHTable 3. Sampling results (
- Page 78 and 79:
RESEARCHpresence of Legionella spp.
- Page 80 and 81:
Seroprevalence for Hepatitis Eand O
- Page 82 and 83:
RESEARCHTable 1. Description of stu
- Page 84 and 85:
RESEARCHTable 3. Crude and adjusted
- Page 86 and 87: RESEARCHrates by gender or HIV stat
- Page 88 and 89: RESEARCH25. Taha TE, Kumwenda N, Ka
- Page 90 and 91: POLICY REVIEWDutch Consensus Guidel
- Page 92 and 93: POLICY REVIEWTable 3. Comparison of
- Page 94 and 95: POLICY REVIEW6. Botelho-Nevers E, F
- Page 96 and 97: DISPATCHESFigure 1. Phylogenetic tr
- Page 98 and 99: DISPATCHESSevere Pediatric Adenovir
- Page 100 and 101: DISPATCHESTable 1. Demographics and
- Page 102 and 103: DISPATCHES13. Kim YJ, Hong JY, Lee
- Page 104 and 105: DISPATCHESTable. Alignment of resid
- Page 106 and 107: DISPATCHESFigure 2. Interaction of
- Page 108 and 109: DISPATCHESSchmallenberg Virus Recur
- Page 110 and 111: DISPATCHESFigure 2. Detection of Sc
- Page 112 and 113: DISPATCHESFigure 1. Histopathologic
- Page 114: DISPATCHESFigure 2. Detection of fo
- Page 117 and 118: Influenza Virus Strains in the Amer
- Page 119 and 120: Novel Arenavirus Isolates from Nama
- Page 121 and 122: Novel Arenaviruses, Southern Africa
- Page 123 and 124: Readability of Ebola Informationon
- Page 125 and 126: Readability of Ebola Information on
- Page 127 and 128: Patients under investigation for ME
- Page 129 and 130: Patients under investigation for ME
- Page 131 and 132: Wildlife Reservoir for Hepatitis E
- Page 133 and 134: Asymptomatic Malaria and Other Infe
- Page 135: Asymptomatic Malaria in Children fr
- Page 139 and 140: Bufavirus in Wild Shrews and Nonhum
- Page 141 and 142: Range Expansion for Rat Lungworm in
- Page 143 and 144: Slow Clearance of Plasmodium falcip
- Page 145 and 146: Slow Clearance of Plasmodium falcip
- Page 147 and 148: Gastroenteritis Caused by Norovirus
- Page 149 and 150: Ebola Virus Stability on Surfaces a
- Page 151 and 152: Ebola Virus Stability on Surfaces a
- Page 153 and 154: Outbreak of Ciprofloxacin-Resistant
- Page 155 and 156: Outbreak of S. sonnei, South KoreaT
- Page 157 and 158: Rapidly Expanding Range of Highly P
- Page 159 and 160: Cluster of Ebola Virus Disease, Bon
- Page 161 and 162: Cluster of Ebola Virus Disease, Lib
- Page 163 and 164: ANOTHER DIMENSIONThe Past Is Never
- Page 165 and 166: Measles Epidemic, Boston, Massachus
- Page 167 and 168: LETTERSInfluenza A(H5N6)Virus Reass
- Page 169 and 170: LETTERSsystem (8 kb-span paired-end
- Page 171 and 172: LETTERS3. Van Hong N, Amambua-Ngwa
- Page 173 and 174: LETTERSTable. Prevalence of Bartone
- Page 175 and 176: LETTERSavian influenza A(H5N1) viru
- Page 177 and 178: LETTERSprovinces and a total of 200
- Page 179 and 180: LETTERS7. Manian FA. Bloodstream in
- Page 181 and 182: LETTERSforward projections. N Engl
- Page 183 and 184: LETTERS3. Guindon S, Gascuel OA. Si
- Page 185 and 186: BOOKS AND MEDIAin the port cities o
- Page 187 and 188:
ABOUT THE COVERNorth was not intere
- Page 189 and 190:
Earning CME CreditTo obtain credit,
- Page 191:
Emerging Infectious Diseases is a p