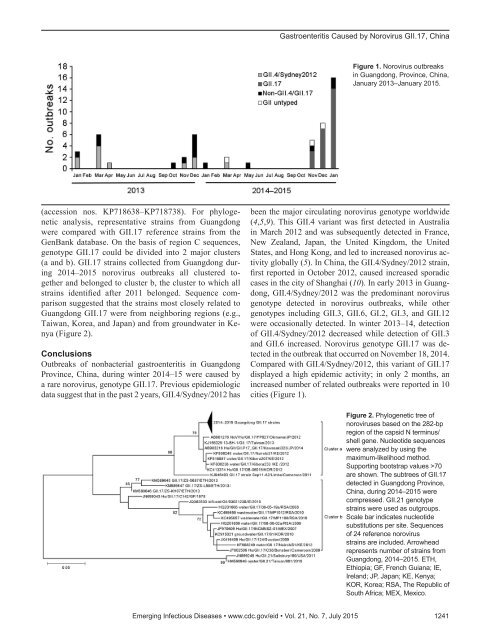
DISPATCHESGastroenteritis Outbreaks Causedby Norovirus GII.17, Guangdong Province,China, 2014–2015Jing Lu, 1 Limei Sun, 1 Lin Fang, Feng Yang,Yanling Mo, Jiaqian Lao, Huanying Zheng,Xiaohua Tan, Hualiang Lin, Shannon Rutherford,Lili Guo, Changwen Ke, Li HuiIn the past decade, the most prevalent norovirus genotypecausing viral gastroenteritis outbreaks worldwide, includingChina, has been GII.4. In winter 2014–15, norovirus outbreaksin Guangdong, China, increased. Sequence analysisindicated that 82% of the outbreaks were caused by anorovirus GII.17 variant.Norovirus infection is a leading cause of nonbacterialgastroenteritis outbreaks in industrialized and developingcountries (1,2). On the basis of amino acid identityin viral protein 1, noroviruses can be divided into atleast 6 genogroups (GI–GVI). GI and GII infect humansand can be further classified into genotypes; at least 9genotypes belong to GI and 22 belong to GII (3). Duringthe past decade, most reported norovirus outbreakswere caused by GII.4 norovirus (4,5). New variants ofGII.4 have emerged approximately every 2–3 years andhave caused norovirus gastroenteritis pandemics globally(6). Since 1999, the major circulating genotype inmainland China has been GII.4, accounting for 64% ofall genotypes detected (7). In winter 2014–15, norovirusoutbreaks in Guangdong Province, China, increased. Sequenceanalyses showed that the major cause of continuousgastroenteritis outbreaks in the region was a rarelyreported norovirus genotype: GII.17.The StudyIn China, according to the National Public Health EmergencyContingency Plan, an outbreak with a cluster of atleast 20 acute gastroenteritis cases (meeting the Kaplan criterion)within 3 days must be reported to Guangdong ProvincialCenter for Disease Control and Prevention. Samplesfrom each outbreak are first tested for norovirus (NorovirusAuthor affiliations: Guangdong Provincial Center for DiseaseControl and Prevention, Guangzhou, China (J. Lu, L. Sun,L. Fang, F. Yang, Y. Mo, J. Lao, H. Zheng, X. Tan, H. Lin, L. Guo,C. Ke, L. Hui); Guangdong Provincial Institution of Public Health,Guangzhou (J. Lu, H. Lin); Centre for Environment and PopulationHealth, Brisbane, Queensland, Australia (S. Rutherford)DOI: http://dx.doi.org/10.3201/eid2107.150226RT-PCR Kit; Shanghai ZJ Bio-Tech Co., Ltd., Shanghai,China) and for intestinal bacteria at the local Centers for DiseaseControl and Prevention. The norovirus-positive specimensare delivered to the Guangdong Provincial Center forDisease Control and Prevention for further genotyping.From each outbreak, 5–10 samples are randomly selectedfor sequencing. Testing with One-Step RT-PCR(QIAGEN, Valencia, CA, USA) is performed with regionC–specific primer, as previously described (8). The positivePCR products are sequenced, and norovirus genotypes aredetermined by using the Norovirus Automated GenotypingTool (http://www.rivm.nl/mpf/norovirus/typingtool) orblastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The majornorovirus genotype causing each outbreak is defined as 1genotype detected in >80% samples from the outbreak.From January 2013 through January 2015, a total of 52norovirus outbreaks were reported and were associated with4,618 clinical cases; of these, 14 outbreaks were associatedwith ≈100 clinical cases. Of the 52 outbreaks, 44 (85%)occurred in schools and colleges, 5 (9.6%) in factories, and3 (5.7%) in kindergartens. In Guangdong Province, norovirusoutbreaks are highly seasonal; most (96%) outbreaksare reported from November through March (Figure 1). Inlate 2014, an increase in the number of norovirus outbreakswas noted. From November 2014 through January 2015, atotal of 29 identified outbreaks were associated with 2,340cases compared with 9 outbreaks and 949 cases the previouswinter (2013–14).Samples from 46 (88%) of the 52 outbreaks were successfullygenotyped. GII norovirus was detected in samplesfrom 96% of the outbreaks. From January 2013 through October2014, the most common genotype found was GII.4/Sydney/2012, which was detected in samples from 48% ofthe outbreaks. Genotype GII.17 was first detected in the cityof Guangzhou in November 2014 and thereafter spread rapidly(online Technical Appendix Figure 1, http://wwwnc.cdc.gov/EID/article/21/7/15-0226-Techapp1.<strong>pdf</strong>). FromNovember 2014 through January 2015, GII.17 norovirusoutbreaks were reported in 10 cities of Guangdong Provinceand represented 83% (24 of 29) of all outbreaks. In contrast,during 2013 and 2014, norovirus outbreaks caused by GII.4/Sydney/2012 were reported in only 5 cities in Guangdong.The nucleotide sequences of the norovirus GII.17strains from Guangdong have been deposited in GenBank1These authors contributed equally to this article.1240 Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015
Gastroenteritis Caused by Norovirus GII.17, ChinaFigure 1. Norovirus outbreaksin Guangdong, Province, China,January 2013–January 2015.(accession nos. KP718638–KP718738). For phylogeneticanalysis, representative strains from Guangdongwere compared with GII.17 reference strains from theGenBank database. On the basis of region C sequences,genotype GII.17 could be divided into 2 major clusters(a and b). GII.17 strains collected from Guangdong during2014–2015 norovirus outbreaks all clustered togetherand belonged to cluster b, the cluster to which allstrains identified after 2011 belonged. Sequence comparisonsuggested that the strains most closely related toGuangdong GII.17 were from neighboring regions (e.g.,Taiwan, Korea, and Japan) and from groundwater in Kenya(Figure 2).ConclusionsOutbreaks of nonbacterial gastroenteritis in GuangdongProvince, China, during winter 2014–15 were caused bya rare norovirus, genotype GII.17. Previous epidemiologicdata suggest that in the past 2 years, GII.4/Sydney/2012 hasbeen the major circulating norovirus genotype worldwide(4,5,9). This GII.4 variant was first detected in Australiain March 2012 and was subsequently detected in France,New Zealand, Japan, the United Kingdom, the UnitedStates, and Hong Kong, and led to increased norovirus activityglobally (5). In China, the GII.4/Sydney/2012 strain,first reported in October 2012, caused increased sporadiccases in the city of Shanghai (10). In early 2013 in Guangdong,GII.4/Sydney/2012 was the predominant norovirusgenotype detected in norovirus outbreaks, while othergenotypes including GII.3, GII.6, GI.2, GI.3, and GII.12were occasionally detected. In winter 2013–14, detectionof GII.4/Sydney/2012 decreased while detection of GII.3and GII.6 increased. Norovirus genotype GII.17 was detectedin the outbreak that occurred on November 18, 2014.Compared with GII.4/Sydney/2012, this variant of GII.17displayed a high epidemic activity; in only 2 months, anincreased number of related outbreaks were reported in 10cities (Figure 1).Figure 2. Phylogenetic tree ofnoroviruses based on the 282-bpregion of the capsid N terminus/shell gene. Nucleotide sequenceswere analyzed by using themaximum-likelihood method.Supporting bootstrap values >70are shown. The subtrees of GII.17detected in Guangdong Province,China, during 2014–2015 werecompressed. GII.21 genotypestrains were used as outgroups.Scale bar indicates nucleotidesubstitutions per site. Sequencesof 24 reference norovirusstrains are included. Arrowheadrepresents number of strains fromGuangdong, 2014–2015. ETH,Ethiopia; GF, French Guiana; IE,Ireland; JP, Japan; KE, Kenya;KOR, Korea; RSA, The Republic ofSouth Africa; MEX, Mexico.Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015 1241
- Page 3 and 4:
July 2015SynopsisOn the CoverMarian
- Page 5 and 6:
1240 Gastroenteritis OutbreaksCause
- Page 7 and 8:
SYNOPSISDisseminated Infections wit
- Page 9 and 10:
Disseminated Infections with Talaro
- Page 11 and 12:
Disseminated Infections with Talaro
- Page 13 and 14:
Macacine Herpesvirus 1 inLong-Taile
- Page 15 and 16:
Macacine Herpesvirus 1 in Macaques,
- Page 17 and 18:
Macacine Herpesvirus 1 in Macaques,
- Page 19:
Macacine Herpesvirus 1 in Macaques,
- Page 23:
Malaria among Young Infants, Africa
- Page 26 and 27:
RESEARCHFigure 3. Dynamics of 19-kD
- Page 28 and 29:
Transdermal Diagnosis of MalariaUsi
- Page 30 and 31:
RESEARCHFigure 2. A) Acoustic trace
- Page 32 and 33:
RESEARCHof malaria-infected mosquit
- Page 34 and 35:
Lack of Transmission amongClose Con
- Page 36 and 37:
RESEARCH(IFA) and microneutralizati
- Page 38 and 39:
RESEARCHoropharyngeal, and serum sa
- Page 40 and 41:
RESEARCH6. Assiri A, McGeer A, Perl
- Page 42 and 43:
RESEARCHadvanced genomic sequencing
- Page 44 and 45:
RESEARCHTable 2. Next-generation se
- Page 46 and 47:
RESEARCHTable 3. Mutation analysis
- Page 48 and 49:
RESEARCHReferences1. Baize S, Panne
- Page 50 and 51:
Parechovirus Genotype 3 Outbreakamo
- Page 52 and 53:
RESEARCHFigure 1. Venn diagramshowi
- Page 54 and 55:
RESEARCHTable 2. HPeV testing of sp
- Page 56 and 57:
RESEARCHFigure 5. Distribution of h
- Page 58 and 59:
RESEARCHReferences1. Selvarangan R,
- Page 60 and 61:
RESEARCHthe left lobe was sampled b
- Page 62 and 63:
RESEARCHTable 2. Middle East respir
- Page 64 and 65:
RESEARCHseroprevalence in domestic
- Page 66 and 67:
RESEARCHmeasure their current surve
- Page 68 and 69:
RESEARCHTable 2. States with labora
- Page 70 and 71:
RESEARCHFigure 2. Comparison of sur
- Page 72 and 73:
RESEARCH9. Centers for Disease Cont
- Page 74 and 75:
RESEARCHthe analyses. Cases in pers
- Page 76 and 77:
RESEARCHTable 3. Sampling results (
- Page 78 and 79:
RESEARCHpresence of Legionella spp.
- Page 80 and 81:
Seroprevalence for Hepatitis Eand O
- Page 82 and 83:
RESEARCHTable 1. Description of stu
- Page 84 and 85:
RESEARCHTable 3. Crude and adjusted
- Page 86 and 87:
RESEARCHrates by gender or HIV stat
- Page 88 and 89:
RESEARCH25. Taha TE, Kumwenda N, Ka
- Page 90 and 91:
POLICY REVIEWDutch Consensus Guidel
- Page 92 and 93:
POLICY REVIEWTable 3. Comparison of
- Page 94 and 95:
POLICY REVIEW6. Botelho-Nevers E, F
- Page 96 and 97: DISPATCHESFigure 1. Phylogenetic tr
- Page 98 and 99: DISPATCHESSevere Pediatric Adenovir
- Page 100 and 101: DISPATCHESTable 1. Demographics and
- Page 102 and 103: DISPATCHES13. Kim YJ, Hong JY, Lee
- Page 104 and 105: DISPATCHESTable. Alignment of resid
- Page 106 and 107: DISPATCHESFigure 2. Interaction of
- Page 108 and 109: DISPATCHESSchmallenberg Virus Recur
- Page 110 and 111: DISPATCHESFigure 2. Detection of Sc
- Page 112 and 113: DISPATCHESFigure 1. Histopathologic
- Page 114: DISPATCHESFigure 2. Detection of fo
- Page 117 and 118: Influenza Virus Strains in the Amer
- Page 119 and 120: Novel Arenavirus Isolates from Nama
- Page 121 and 122: Novel Arenaviruses, Southern Africa
- Page 123 and 124: Readability of Ebola Informationon
- Page 125 and 126: Readability of Ebola Information on
- Page 127 and 128: Patients under investigation for ME
- Page 129 and 130: Patients under investigation for ME
- Page 131 and 132: Wildlife Reservoir for Hepatitis E
- Page 133 and 134: Asymptomatic Malaria and Other Infe
- Page 135 and 136: Asymptomatic Malaria in Children fr
- Page 137 and 138: Bufavirus in Wild Shrews and Nonhum
- Page 139 and 140: Bufavirus in Wild Shrews and Nonhum
- Page 141 and 142: Range Expansion for Rat Lungworm in
- Page 143 and 144: Slow Clearance of Plasmodium falcip
- Page 145: Slow Clearance of Plasmodium falcip
- Page 149 and 150: Ebola Virus Stability on Surfaces a
- Page 151 and 152: Ebola Virus Stability on Surfaces a
- Page 153 and 154: Outbreak of Ciprofloxacin-Resistant
- Page 155 and 156: Outbreak of S. sonnei, South KoreaT
- Page 157 and 158: Rapidly Expanding Range of Highly P
- Page 159 and 160: Cluster of Ebola Virus Disease, Bon
- Page 161 and 162: Cluster of Ebola Virus Disease, Lib
- Page 163 and 164: ANOTHER DIMENSIONThe Past Is Never
- Page 165 and 166: Measles Epidemic, Boston, Massachus
- Page 167 and 168: LETTERSInfluenza A(H5N6)Virus Reass
- Page 169 and 170: LETTERSsystem (8 kb-span paired-end
- Page 171 and 172: LETTERS3. Van Hong N, Amambua-Ngwa
- Page 173 and 174: LETTERSTable. Prevalence of Bartone
- Page 175 and 176: LETTERSavian influenza A(H5N1) viru
- Page 177 and 178: LETTERSprovinces and a total of 200
- Page 179 and 180: LETTERS7. Manian FA. Bloodstream in
- Page 181 and 182: LETTERSforward projections. N Engl
- Page 183 and 184: LETTERS3. Guindon S, Gascuel OA. Si
- Page 185 and 186: BOOKS AND MEDIAin the port cities o
- Page 187 and 188: ABOUT THE COVERNorth was not intere
- Page 189 and 190: Earning CME CreditTo obtain credit,
- Page 191: Emerging Infectious Diseases is a p