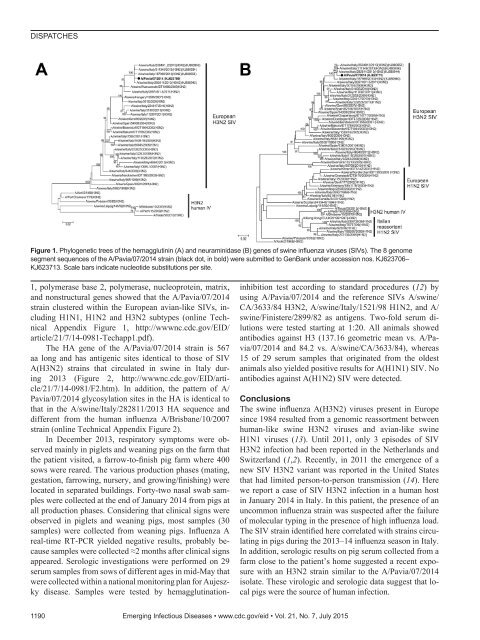
DISPATCHESFigure 1. Phylogenetic trees of the hemagglutinin (A) and neuraminidase (B) genes of swine influenza viruses (SIVs). The 8 genomesegment sequences of the A/Pavia/07/2014 strain (black dot, in bold) were submitted to GenBank under accession nos. KJ623706–KJ623713. Scale bars indicate nucleotide substitutions per site.1, polymerase base 2, polymerase, nucleoprotein, matrix,and nonstructural genes showed that the A/Pavia/07/2014strain clustered within the European avian-like SIVs, includingH1N1, H1N2 and H3N2 subtypes (online TechnicalAppendix Figure 1, http://wwwnc.cdc.gov/EID/article/21/7/14-0981-Techapp1.<strong>pdf</strong>).The HA gene of the A/Pavia/07/2014 strain is 567aa long and has antigenic sites identical to those of SIVA(H3N2) strains that circulated in swine in Italy during2013 (Figure 2, http://wwwnc.cdc.gov/EID/article/21/7/14-0981/F2.htm).In addition, the pattern of A/Pavia/07/2014 glycosylation sites in the HA is identical tothat in the A/swine/Italy/282811/2013 HA sequence anddifferent from the human influenza A/Brisbane/10/2007strain (online Technical Appendix Figure 2).In December 2013, respiratory symptoms were observedmainly in piglets and weaning pigs on the farm thatthe patient visited, a farrow-to-finish pig farm where 400sows were reared. The various production phases (mating,gestation, farrowing, nursery, and growing/finishing) werelocated in separated buildings. Forty-two nasal swab sampleswere collected at the end of January 2014 from pigs atall production phases. Considering that clinical signs wereobserved in piglets and weaning pigs, most samples (30samples) were collected from weaning pigs. Influenza Areal-time RT-PCR yielded negative results, probably becausesamples were collected ≈2 months after clinical signsappeared. Serologic investigations were performed on 29serum samples from sows of different ages in mid-May thatwere collected within a national monitoring plan for Aujeszkydisease. Samples were tested by hemagglutination-inhibition test according to standard procedures (12) byusing A/Pavia/07/2014 and the reference SIVs A/swine/CA/3633/84 H3N2, A/swine/Italy/1521/98 H1N2, and A/swine/Finistere/2899/82 as antigens. Two-fold serum dilutionswere tested starting at 1:20. All animals showedantibodies against H3 (137.16 geometric mean vs. A/Pavia/07/2014and 84.2 vs. A/swine/CA/3633/84), whereas15 of 29 serum samples that originated from the oldestanimals also yielded positive results for A(H1N1) SIV. Noantibodies against A(H1N2) SIV were detected.ConclusionsThe swine influenza A(H3N2) viruses present in Europesince 1984 resulted from a genomic reassortment betweenhuman-like swine H3N2 viruses and avian-like swineH1N1 viruses (13). Until 2011, only 3 episodes of SIVH3N2 infection had been reported in the Netherlands andSwitzerland (1,2). Recently, in 2011 the emergence of anew SIV H3N2 variant was reported in the United Statesthat had limited person-to-person transmission (14). Herewe report a case of SIV H3N2 infection in a human hostin January 2014 in Italy. In this patient, the presence of anuncommon influenza strain was suspected after the failureof molecular typing in the presence of high influenza load.The SIV strain identified here correlated with strains circulatingin pigs during the 2013–14 influenza season in Italy.In addition, serologic results on pig serum collected from afarm close to the patient’s home suggested a recent exposurewith an H3N2 strain similar to the A/Pavia/07/2014isolate. These virologic and serologic data suggest that localpigs were the source of human infection.1190 Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015
Swine Influenza, ItalyIn agreement with previous observations (1,2), the EuropeanH3N2 swine viruses seem to cause a benign diseasewith mild influenza-like symptoms in humans. In addition,the SIV strain we identified was found only in an immunocompromisedpatient. The uncomplicated clinical coursemight not be uncommon in immunocompromised patients.Indeed, in 2 epidemiologically correlated immunocompromisedpatients, the emergence of human influenza H3N2-resistant strains was associated with opposite clinical outcomes:1 patient had mild upper respiratory syndrome; theother died of severe acute respiratory distress syndrome(15). On the other hand, on the basis of the observationthat none of the patient’s family members and co-workersshowed respiratory infection, we can hypothesize that immuneimpairment of the patient could have favored thezoonotic transmission of the SIV strain. Surveillance ofcirculating SIVs and monitoring of occupationally exposedworkers are 2 important tools to prevent spread of potentialpandemic viruses.Additional members of the Influenza Study Group who contributeddata: Alessia Griello, Marta Premoli, Franscesca Rovida,Bianca Mariani (SS Virologia Molecolare, SC Microbiologia eVirologia, Fondazione IRCCS Policlinico San Matteo, Pavia,Italy); Francesca Manola Adella (Dipartimento di Virologia,Istituto Zooprofilattico Sperimentale della Lombardia ed EmiliaRomagna, Brescia, Italy); Anna Maria Belloni (Azienda SanitariaLocale); Maria Gramegna, Liliana Coppola, AlessandraPiatti, Laura Gemma Brenzoni (DG Sanità, Regione Lombardia,Milan, Italy); Mario Luini (Organizzazione Mondiale per laSalute degli Animali, Laboratorio di riferimento per l’InfluenzaSuina, Istituto Zooprofilattico Sperimentale della Lombardiaed Emilia Romagna, Parma, Italy); Emanuela Foni and LauraBaioni (Istituto Zooprofilattico Sperimentale della Lombardia edEmilia Romagna, Parma, Italy)AcknowledgmentsWe thank the Direzione Generale Sanità, Regione Lombardia;the physicians and veterinarians of the Azienda Sanitaria Localeinvolved in case management; and all the collaborators in thecase definition. We thank Daniela Sartori for manuscript editingand Laurene Kelly for English revision.This study was supported by grants from the Ministero della Salute,Fondazione IRCCS Policlinico San Matteo, Ricerca Corrente(grant no. 80622), Progetto Cariplo 2011-0517, Milan, Italy, andby a grant from the Ministero della Salute, IZSLER PRC2012002.Dr. Piralla is a clinical virologist at the Molecular Virology Unitof the Fondazione IRCCS Policlinico San Matteo in Pavia, Italy.His main research interests include molecular epidemiology of respiratoryviruses, the study of virus evolution and interaction withthe host, and design of next-generation sequencing protocols tostudy virus evolution and new pathogen discovery.References1. Zell R, Scholtissek C, Ludwig S. Genetics, evolution, and the zoonoticcapacity of European swine influenza viruses. Curr Top MicrobiolImmunol. 2013;370:29–55. http://dx.doi.org/10.1007/82_2012_2672. Myers KP, Olsen CW, Gray GC. Cases of swine influenza inhumans: a review of the literature. Clin Infect Dis. 2007;44:1084–8. http://dx.doi.org/10.1086/5128133. Krumbholz A, Lange J, Dürrwald R, Walther M, Muller TH,Kuhnel D, et al. Prevalence of antibodies to European porcineinfluenza viruses in humans living in high pig density areas ofGermany. Med Microbiol Immunol (Berl). 2014;203:13–24.http://dx.doi.org/10.1007/s00430-013-0309-y4. Gerloff NA, Kremer JR, Charpentier E, Sausy A, Olinger CM,Weicherding P, et al. Swine influenza virus antibodies in humans,western Europe, 2009. Emerg Infect Dis. 2011;17:403–11.http://dx.doi.org/10.3201/eid1703.1008515. Piralla A, Baldanti F, Gerna G. Phylogenetic patterns of humanrespiratory picornavirus species, including the newly identifiedgroup C rhinoviruses, during a 1-year surveillance of a hospitalizedpatient population in Italy. J Clin Microbiol. 2011;49:373–6.http://dx.doi.org/10.1128/JCM.01814-106. World Health Organization. CDC protocol of realtime RTPCRfor influenza A(H1N1). 2009 Oct 6 [cited 2009 Dec 15].http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.<strong>pdf</strong>7. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primerset for the full-length amplification of all influenza A viruses. ArchVirol. 2001;146:2275–89. http://dx.doi.org/10.1007/s0070501700028. Lycett SJ, Baillie G, Coulter E, Bhatt S, Kellam P, McCauley JW,et al. Estimating reassortment rates in co-circulating Eurasianswine influenza viruses. J Gen Virol. 2012;93:2326–36.http://dx.doi.org/10.1099/vir.0.044503-09. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W,Gascuel O. New algorithms and methods to estimate maximum-likelihoodphylogenies: assessing the performance of PhyML 3.0. SystBiol. 2010;59:307–21. http://dx.doi.org/10.1093/sysbio/syq01010. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.MEGA5: molecular evolutionary genetics analysis using maximumlikelihood, evolutionary distance, and maximum parsimonymethods. Mol Biol Evol. 2011;28:2731–9. http://dx.doi.org/10.1093/molbev/msr12111 Tharakaraman K, Raman R, Stebbins NW, Viswanathan K,Sasisekharan V, Sasisekharan R. Antigenically intact hemagglutinin incirculating avian and swine influenza viruses and potential for H3N2pandemic. Sci Rep. 2013;3:1822. http://dx.doi.org/10.1038/srep0182212. World Organisation for Animal Health. Manual of diagnostic testsand vaccines for terrestrial animals. Chapter 2.08.08. Swine influenza[cited 2014 Jun 6]. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.08.08_SWINE_INFLUENZA.<strong>pdf</strong>13. Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y,Webster RG. Genetic reassortant between avian and humaninfluenza a viruses in Italian pigs. Virology. 1993;193:503–6.http://dx.doi.org/10.1006/viro.1993.115514. Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M,et al. Human infections with influenza A(H3N2) variant virusin the United States, 2011–2012. Clin Infect Dis. 2013;57(Suppl 1):S4–11. http://dx.doi.org/10.1093/cid/cit27215. Piralla A, Gozalo-Margüello M, Fiorina L, Rovida F, Muzzi A,Colombo AA, et al. Different drug-resistant influenza A(H3N2)variants in two immunocompromised patients treated with oseltamivirduring the 2011–2012 influenza season in Italy. J Clin Virol.2013;58:132–7. http://dx.doi.org/10.1016/j.jcv.2013.06.003Address for correspondence: Fausto Baldanti, SS Virologia Molecolare,SC Microbiologia e Virologia, Fondazione IRCCS Policlinico San Matteo,via Taramelli 5, 27100 Pavia, Italy; email: f.baldanti@smatteo.pv.itEmerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015 1191
- Page 3 and 4:
July 2015SynopsisOn the CoverMarian
- Page 5 and 6:
1240 Gastroenteritis OutbreaksCause
- Page 7 and 8:
SYNOPSISDisseminated Infections wit
- Page 9 and 10:
Disseminated Infections with Talaro
- Page 11 and 12:
Disseminated Infections with Talaro
- Page 13 and 14:
Macacine Herpesvirus 1 inLong-Taile
- Page 15 and 16:
Macacine Herpesvirus 1 in Macaques,
- Page 17 and 18:
Macacine Herpesvirus 1 in Macaques,
- Page 19:
Macacine Herpesvirus 1 in Macaques,
- Page 23:
Malaria among Young Infants, Africa
- Page 26 and 27:
RESEARCHFigure 3. Dynamics of 19-kD
- Page 28 and 29:
Transdermal Diagnosis of MalariaUsi
- Page 30 and 31:
RESEARCHFigure 2. A) Acoustic trace
- Page 32 and 33:
RESEARCHof malaria-infected mosquit
- Page 34 and 35:
Lack of Transmission amongClose Con
- Page 36 and 37:
RESEARCH(IFA) and microneutralizati
- Page 38 and 39:
RESEARCHoropharyngeal, and serum sa
- Page 40 and 41:
RESEARCH6. Assiri A, McGeer A, Perl
- Page 42 and 43:
RESEARCHadvanced genomic sequencing
- Page 44 and 45:
RESEARCHTable 2. Next-generation se
- Page 46 and 47: RESEARCHTable 3. Mutation analysis
- Page 48 and 49: RESEARCHReferences1. Baize S, Panne
- Page 50 and 51: Parechovirus Genotype 3 Outbreakamo
- Page 52 and 53: RESEARCHFigure 1. Venn diagramshowi
- Page 54 and 55: RESEARCHTable 2. HPeV testing of sp
- Page 56 and 57: RESEARCHFigure 5. Distribution of h
- Page 58 and 59: RESEARCHReferences1. Selvarangan R,
- Page 60 and 61: RESEARCHthe left lobe was sampled b
- Page 62 and 63: RESEARCHTable 2. Middle East respir
- Page 64 and 65: RESEARCHseroprevalence in domestic
- Page 66 and 67: RESEARCHmeasure their current surve
- Page 68 and 69: RESEARCHTable 2. States with labora
- Page 70 and 71: RESEARCHFigure 2. Comparison of sur
- Page 72 and 73: RESEARCH9. Centers for Disease Cont
- Page 74 and 75: RESEARCHthe analyses. Cases in pers
- Page 76 and 77: RESEARCHTable 3. Sampling results (
- Page 78 and 79: RESEARCHpresence of Legionella spp.
- Page 80 and 81: Seroprevalence for Hepatitis Eand O
- Page 82 and 83: RESEARCHTable 1. Description of stu
- Page 84 and 85: RESEARCHTable 3. Crude and adjusted
- Page 86 and 87: RESEARCHrates by gender or HIV stat
- Page 88 and 89: RESEARCH25. Taha TE, Kumwenda N, Ka
- Page 90 and 91: POLICY REVIEWDutch Consensus Guidel
- Page 92 and 93: POLICY REVIEWTable 3. Comparison of
- Page 94 and 95: POLICY REVIEW6. Botelho-Nevers E, F
- Page 98 and 99: DISPATCHESSevere Pediatric Adenovir
- Page 100 and 101: DISPATCHESTable 1. Demographics and
- Page 102 and 103: DISPATCHES13. Kim YJ, Hong JY, Lee
- Page 104 and 105: DISPATCHESTable. Alignment of resid
- Page 106 and 107: DISPATCHESFigure 2. Interaction of
- Page 108 and 109: DISPATCHESSchmallenberg Virus Recur
- Page 110 and 111: DISPATCHESFigure 2. Detection of Sc
- Page 112 and 113: DISPATCHESFigure 1. Histopathologic
- Page 114: DISPATCHESFigure 2. Detection of fo
- Page 117 and 118: Influenza Virus Strains in the Amer
- Page 119 and 120: Novel Arenavirus Isolates from Nama
- Page 121 and 122: Novel Arenaviruses, Southern Africa
- Page 123 and 124: Readability of Ebola Informationon
- Page 125 and 126: Readability of Ebola Information on
- Page 127 and 128: Patients under investigation for ME
- Page 129 and 130: Patients under investigation for ME
- Page 131 and 132: Wildlife Reservoir for Hepatitis E
- Page 133 and 134: Asymptomatic Malaria and Other Infe
- Page 135 and 136: Asymptomatic Malaria in Children fr
- Page 137 and 138: Bufavirus in Wild Shrews and Nonhum
- Page 139 and 140: Bufavirus in Wild Shrews and Nonhum
- Page 141 and 142: Range Expansion for Rat Lungworm in
- Page 143 and 144: Slow Clearance of Plasmodium falcip
- Page 145 and 146: Slow Clearance of Plasmodium falcip
- Page 147 and 148:
Gastroenteritis Caused by Norovirus
- Page 149 and 150:
Ebola Virus Stability on Surfaces a
- Page 151 and 152:
Ebola Virus Stability on Surfaces a
- Page 153 and 154:
Outbreak of Ciprofloxacin-Resistant
- Page 155 and 156:
Outbreak of S. sonnei, South KoreaT
- Page 157 and 158:
Rapidly Expanding Range of Highly P
- Page 159 and 160:
Cluster of Ebola Virus Disease, Bon
- Page 161 and 162:
Cluster of Ebola Virus Disease, Lib
- Page 163 and 164:
ANOTHER DIMENSIONThe Past Is Never
- Page 165 and 166:
Measles Epidemic, Boston, Massachus
- Page 167 and 168:
LETTERSInfluenza A(H5N6)Virus Reass
- Page 169 and 170:
LETTERSsystem (8 kb-span paired-end
- Page 171 and 172:
LETTERS3. Van Hong N, Amambua-Ngwa
- Page 173 and 174:
LETTERSTable. Prevalence of Bartone
- Page 175 and 176:
LETTERSavian influenza A(H5N1) viru
- Page 177 and 178:
LETTERSprovinces and a total of 200
- Page 179 and 180:
LETTERS7. Manian FA. Bloodstream in
- Page 181 and 182:
LETTERSforward projections. N Engl
- Page 183 and 184:
LETTERS3. Guindon S, Gascuel OA. Si
- Page 185 and 186:
BOOKS AND MEDIAin the port cities o
- Page 187 and 188:
ABOUT THE COVERNorth was not intere
- Page 189 and 190:
Earning CME CreditTo obtain credit,
- Page 191:
Emerging Infectious Diseases is a p