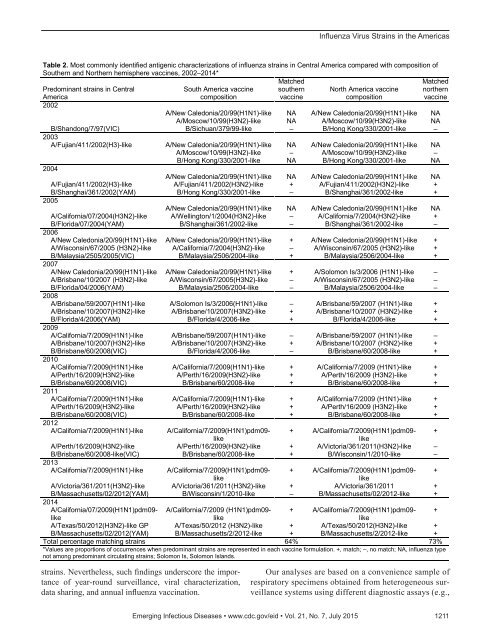
DISPATCHESin Central America and 12 (92%, 95% CI 78%–107%)of 13 subsequent seasons in North America (Table 1; onlineTechnical Appendix, http://wwwnc.cdc.gov/EID/article/21/7/14-0788-Techapp1.xlxs). Similarly, A(H3N2)virus strains in South America predominated in all 11 (92%,95% CI 76%–107%) of 12 subsequent seasons in CentralAmerica and 10 (71%, 95% CI 48%–95%) of 14 subsequentseasons in North America. Predominant influenza B virusstrains in South America only predominated in 8 (67%, 95%CI 40%–93%) of 12 subsequent seasons in Central Americaand 8 (57%, 95% CI 31%–83%) of 14 subsequent seasonsin North America. Virus strains in South America during 1season typically did not predominate in subsequent seasonsin South America (54%, 95% CI 38%–70%).The predominant A(H1N1) virus strains in NorthAmerica predominated in 7 (78%, 95% CI 51%–105%) of 9subsequent seasons in Central America and 10 (83%, 95%CI 62%–104%) of 12 subsequent seasons in South America.A(H3N2) virus strains in North America predominatedin 8 (67%, 95% CI 40%–93%) of 12 subsequent seasonsin Central America and 10 (77%, 95% CI 54%–100%) of13 subsequent seasons in South America. Influenza B virusstrains in North America predominated in 9 (75%, 95% CI51%–100%) of 12 subsequent seasons in Central Americaand 7 (54%, 95% CI 27%–81%) of 13 subsequent seasonsin South America. Virus strains that predominated in NorthAmerica during 1 season were less likely to predominatein the subsequent season in North America (62%, 95% CI46%–77%).At least 1 component of the Southern Hemisphere vaccinecomposition recommendations matched a predominantantigenic characterization in South America in 13 (93%,95% CI 79%–106% of 14 influenza seasons that occurredduring 2001–2014, and at least 1 component of the NorthernHemisphere vaccine composition recommendationsmatched a predominant antigenic characterization in NorthAmerica in all 14 influenza seasons that occurred during2001–2014. Of 33 predominant antigenic virus strainsidentified in Central America during 2002–2014, 21 (64%,95% CI 47%–80%) matched the Southern Hemisphere recommendationsand 24 (73%, 95% CI 58%–88%) matchedthe Northern Hemisphere recommendations (Table 2).ConclusionsOur findings suggest that virus strains identified duringinfluenza epidemics in South America typically becamepredominant in subsequent epidemics in Central and NorthAmerica. Almost as frequently, virus strains identifiedduring epidemics in North America became predominantin the subsequent Central and South America epidemics.Although strain selection for 1 hemisphere’s vaccine formulationtypically occurs before influenza activity is widespreadin the opposite hemisphere, health officials have anopportunity to anticipate which influenza virus strains maypredominate by observing activity in other subregions. Forexample, influenza A(H1N1)pdm09 virus predominated inBrazil during 2013 (8) and became predominant in NorthAmerica during 2013–2014. Health officials identifyinginfluenza B virus strains in 1 hemisphere would have correctlypredicted the predominant influenza B virus strainsin the opposite hemisphere only half of the time unless theyhad also examined other co-circulating influenza B virusTable 1. Most commonly identified antigenic characterizations of influenza strains in the Americas during 2001–2014*†Influenza A(H1N1) virus Influenza A(H3N2) virus Influenza B virusYear South Central North South Central North South Central North2001 A NA A A NA A A NA B‡2002 A NA A A NA A B B B2003 A NA A A B B‡ B NA C‡2004 NA NA A B B C‡ C C C2005 A NA A C C C D‡ E‡ D2006 A A A D‡ D D E D/E D2007 A A B‡ E‡ E E F‡ F F2008 C‡ C C E E E F F G‡2009 D‡ D D E E F‡ F G G2010 D D D F F F G G G2011 D D D F F F G G G2012 D D D F F F H‡ G H2013 D D D G‡ G H‡ G I‡ I2014 D D D H H H I I I*Data from Canada, Mexico, and the United States were aggregated to represent North America; data from Belize, Costa Rica, El Salvador, Guatemala,Honduras, Nicaragua, and Panama to represent Central America; and data from Argentina, Brazil, Chile, Paraguay, and Uruguay to represent SouthAmerica.†For influenza A(H1N1) virus, A, A/New Caledonia/20/99(H1N1); B, A/Solomon Islands/03/2006(H1N1); C, A/Brisbane/59/2007(H1N1); D,A/California/07/2009-(H1N1)pdm09. For influenza A(H3N2) virus, A, A/Panama/2007/99(H3N2); B, A/Fujian/411/2002(H3); C,A/California/07/2004(H3N2); D, A/Wisconsin/67/2005(H3N2); E, A/Brisbane/10/2007(H3N2); F, A/Perth/16/2009(H3N2); G, A/Victoria/361/2011(H3N2);H, A/Texas/50/2012(H3N2). For influenza B virus, A, B/Sichuan/379/99(YAM); B, B/Shandong/7/97(VIC); C, B/Shanghai/361/2002(YAM); D,B/Malaysia/2505/2005(VIC); E, B/Florida/07/2004(YAM); F, B/Florida/04/2006(YAM); G, B/Brisbane/60/2008(VIC); H, B/Wisconsin/01/2010(YAM) I,B/Massachusetts/02/2012(YAM). NA, not applicable.‡Newly identified strain (8 in South America, 2 in Central America, and 7 in North America).1210 Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015
Influenza Virus Strains in the AmericasTable 2. Most commonly identified antigenic characterizations of influenza strains in Central America compared with composition ofSouthern and Northern hemisphere vaccines, 2002–2014*MatchedSouth America vaccine southern North America vaccinecompositionvaccinecompositionMatchednorthernvaccinePredominant strains in CentralAmerica2002A/New Caledonia/20/99(H1N1)-like NA A/New Caledonia/20/99(H1N1)-like NAA/Moscow/10/99(H3N2)-like NA A/Moscow/10/99(H3N2)-like NAB/Shandong/7/97(VIC) B/Sichuan/379/99-like – B/Hong Kong/330/2001-like –2003A/Fujian/411/2002(H3)-like A/New Caledonia/20/99(H1N1)-like NA A/New Caledonia/20/99(H1N1)-like NAA/Moscow/10/99(H3N2)-like – A/Moscow/10/99(H3N2)-like –B/Hong Kong/330/2001-like NA B/Hong Kong/330/2001-like NA2004A/New Caledonia/20/99(H1N1)-like NA A/New Caledonia/20/99(H1N1)-like NAA/Fujian/411/2002(H3)-like A/Fujian/411/2002(H3N2)-like + A/Fujian/411/2002(H3N2)-like +B/Shanghai/361/2002(YAM) B/Hong Kong/330/2001-like – B/Shanghai/361/2002-like +2005A/New Caledonia/20/99(H1N1)-like NA A/New Caledonia/20/99(H1N1)-like NAA/California/07/2004(H3N2)-like A/Wellington/1/2004(H3N2)-like – A/California/7/2004(H3N2)-like +B/Florida/07/2004(YAM) B/Shanghai/361/2002-like – B/Shanghai/361/2002-like –2006A/New Caledonia/20/99(H1N1)-like A/New Caledonia/20/99(H1N1)-like + A/New Caledonia/20/99(H1N1)-like +A/Wisconsin/67/2005 (H3N2)-like A/California/7/2004(H3N2)-like – A/Wisconsin/67/2005 (H3N2)-like +B/Malaysia/2505/2005(VIC) B/Malaysia/2506/2004-like + B/Malaysia/2506/2004-like +2007A/New Caledonia/20/99(H1N1)-like A/New Caledonia/20/99(H1N1)-like + A/Solomon Is/3/2006 (H1N1)-like –A/Brisbane/10/2007 (H3N2)-like A/Wisconsin/67/2005(H3N2)-like – A/Wisconsin/67/2005 (H3N2)-like –B/Florida/04/2006(YAM) B/Malaysia/2506/2004-like – B/Malaysia/2506/2004-like –2008A/Brisbane/59/2007(H1N1)-like A/Solomon Is/3/2006(H1N1)-like – A/Brisbane/59/2007 (H1N1)-like +A/Brisbane/10/2007(H3N2)-like A/Brisbane/10/2007(H3N2)-like + A/Brisbane/10/2007 (H3N2)-like +B/Florida/4/2006(YAM) B/Florida/4/2006-like + B/Florida/4/2006-like +2009A/California/7/2009(H1N1)-like A/Brisbane/59/2007(H1N1)-like – A/Brisbane/59/2007 (H1N1)-like –A/Brisbane/10/2007(H3N2)-like A/Brisbane/10/2007(H3N2)-like + A/Brisbane/10/2007 (H3N2)-like +B/Brisbane/60/2008(VIC) B/Florida/4/2006-like – B/Brisbane/60/2008-like +2010A/California/7/2009(H1N1)-like A/California/7/2009(H1N1)-like + A/California/7/2009 (H1N1)-like +A/Perth/16/2009(H3N2)-like A/Perth/16/2009(H3N2)-like + A/Perth/16/2009 (H3N2)-like +B/Brisbane/60/2008(VIC) B/Brisbane/60/2008-like + B/Brisbane/60/2008-like +2011A/California/7/2009(H1N1)-like A/California/7/2009(H1N1)-like + A/California/7/2009 (H1N1)-like +A/Perth/16/2009(H3N2)-like A/Perth/16/2009(H3N2)-like + A/Perth/16/2009 (H3N2)-like +B/Brisbane/60/2008(VIC) B/Brisbane/60/2008-like + B/Brisbane/60/2008-like +2012A/California/7/2009(H1N1)-likeA/California/7/2009(H1N1)pdm09- + A/California/7/2009(H1N1)pdm09- +likelikeA/Perth/16/2009(H3N2)-like A/Perth/16/2009(H3N2)-like + A/Victoria/361/2011(H3N2)-like –B/Brisbane/60/2008-like(VIC) B/Brisbane/60/2008-like + B/Wisconsin/1/2010-like –2013A/California/7/2009(H1N1)-likeA/California/7/2009(H1N1)pdm09- + A/California/7/2009(H1N1)pdm09- +likelikeA/Victoria/361/2011(H3N2)-like A/Victoria/361/2011(H3N2)-like + A/Victoria/361/2011 +B/Massachusetts/02/2012(YAM) B/Wisconsin/1/2010-like – B/Massachusetts/02/2012-like +2014A/California/07/2009(H1N1)pdm09-likeA/California/7/2009 (H1N1)pdm09- + A/California/7/2009(H1N1)pdm09- +likelikeA/Texas/50/2012(H3N2)-like GP A/Texas/50/2012 (H3N2)-like + A/Texas/50/2012(H3N2)-like +B/Massachusetts/02/2012(YAM) B/Massachusetts/2/2012-like + B/Massachusetts/2/2012-like +Total percentage matching strains 64% 73%*Values are proportions of occurrences when predominant strains are represented in each vaccine formulation. +, match; –, no match; NA, influenza typenot among predominant circulating strains; Solomon Is, Solomon Islands.strains. Nevertheless, such findings underscore the importanceof year-round surveillance, viral characterization,data sharing, and annual influenza vaccination.Our analyses are based on a convenience sample ofrespiratory specimens obtained from heterogeneous surveillancesystems using different diagnostic assays (e.g.,Emerging Infectious Diseases • www.cdc.gov/eid • Vol. 21, No. 7, July 2015 1211
- Page 3 and 4:
July 2015SynopsisOn the CoverMarian
- Page 5 and 6:
1240 Gastroenteritis OutbreaksCause
- Page 7 and 8:
SYNOPSISDisseminated Infections wit
- Page 9 and 10:
Disseminated Infections with Talaro
- Page 11 and 12:
Disseminated Infections with Talaro
- Page 13 and 14:
Macacine Herpesvirus 1 inLong-Taile
- Page 15 and 16:
Macacine Herpesvirus 1 in Macaques,
- Page 17 and 18:
Macacine Herpesvirus 1 in Macaques,
- Page 19:
Macacine Herpesvirus 1 in Macaques,
- Page 23:
Malaria among Young Infants, Africa
- Page 26 and 27:
RESEARCHFigure 3. Dynamics of 19-kD
- Page 28 and 29:
Transdermal Diagnosis of MalariaUsi
- Page 30 and 31:
RESEARCHFigure 2. A) Acoustic trace
- Page 32 and 33:
RESEARCHof malaria-infected mosquit
- Page 34 and 35:
Lack of Transmission amongClose Con
- Page 36 and 37:
RESEARCH(IFA) and microneutralizati
- Page 38 and 39:
RESEARCHoropharyngeal, and serum sa
- Page 40 and 41:
RESEARCH6. Assiri A, McGeer A, Perl
- Page 42 and 43:
RESEARCHadvanced genomic sequencing
- Page 44 and 45:
RESEARCHTable 2. Next-generation se
- Page 46 and 47:
RESEARCHTable 3. Mutation analysis
- Page 48 and 49:
RESEARCHReferences1. Baize S, Panne
- Page 50 and 51:
Parechovirus Genotype 3 Outbreakamo
- Page 52 and 53:
RESEARCHFigure 1. Venn diagramshowi
- Page 54 and 55:
RESEARCHTable 2. HPeV testing of sp
- Page 56 and 57:
RESEARCHFigure 5. Distribution of h
- Page 58 and 59:
RESEARCHReferences1. Selvarangan R,
- Page 60 and 61:
RESEARCHthe left lobe was sampled b
- Page 62 and 63:
RESEARCHTable 2. Middle East respir
- Page 64 and 65: RESEARCHseroprevalence in domestic
- Page 66 and 67: RESEARCHmeasure their current surve
- Page 68 and 69: RESEARCHTable 2. States with labora
- Page 70 and 71: RESEARCHFigure 2. Comparison of sur
- Page 72 and 73: RESEARCH9. Centers for Disease Cont
- Page 74 and 75: RESEARCHthe analyses. Cases in pers
- Page 76 and 77: RESEARCHTable 3. Sampling results (
- Page 78 and 79: RESEARCHpresence of Legionella spp.
- Page 80 and 81: Seroprevalence for Hepatitis Eand O
- Page 82 and 83: RESEARCHTable 1. Description of stu
- Page 84 and 85: RESEARCHTable 3. Crude and adjusted
- Page 86 and 87: RESEARCHrates by gender or HIV stat
- Page 88 and 89: RESEARCH25. Taha TE, Kumwenda N, Ka
- Page 90 and 91: POLICY REVIEWDutch Consensus Guidel
- Page 92 and 93: POLICY REVIEWTable 3. Comparison of
- Page 94 and 95: POLICY REVIEW6. Botelho-Nevers E, F
- Page 96 and 97: DISPATCHESFigure 1. Phylogenetic tr
- Page 98 and 99: DISPATCHESSevere Pediatric Adenovir
- Page 100 and 101: DISPATCHESTable 1. Demographics and
- Page 102 and 103: DISPATCHES13. Kim YJ, Hong JY, Lee
- Page 104 and 105: DISPATCHESTable. Alignment of resid
- Page 106 and 107: DISPATCHESFigure 2. Interaction of
- Page 108 and 109: DISPATCHESSchmallenberg Virus Recur
- Page 110 and 111: DISPATCHESFigure 2. Detection of Sc
- Page 112 and 113: DISPATCHESFigure 1. Histopathologic
- Page 114: DISPATCHESFigure 2. Detection of fo
- Page 119 and 120: Novel Arenavirus Isolates from Nama
- Page 121 and 122: Novel Arenaviruses, Southern Africa
- Page 123 and 124: Readability of Ebola Informationon
- Page 125 and 126: Readability of Ebola Information on
- Page 127 and 128: Patients under investigation for ME
- Page 129 and 130: Patients under investigation for ME
- Page 131 and 132: Wildlife Reservoir for Hepatitis E
- Page 133 and 134: Asymptomatic Malaria and Other Infe
- Page 135 and 136: Asymptomatic Malaria in Children fr
- Page 137 and 138: Bufavirus in Wild Shrews and Nonhum
- Page 139 and 140: Bufavirus in Wild Shrews and Nonhum
- Page 141 and 142: Range Expansion for Rat Lungworm in
- Page 143 and 144: Slow Clearance of Plasmodium falcip
- Page 145 and 146: Slow Clearance of Plasmodium falcip
- Page 147 and 148: Gastroenteritis Caused by Norovirus
- Page 149 and 150: Ebola Virus Stability on Surfaces a
- Page 151 and 152: Ebola Virus Stability on Surfaces a
- Page 153 and 154: Outbreak of Ciprofloxacin-Resistant
- Page 155 and 156: Outbreak of S. sonnei, South KoreaT
- Page 157 and 158: Rapidly Expanding Range of Highly P
- Page 159 and 160: Cluster of Ebola Virus Disease, Bon
- Page 161 and 162: Cluster of Ebola Virus Disease, Lib
- Page 163 and 164: ANOTHER DIMENSIONThe Past Is Never
- Page 165 and 166:
Measles Epidemic, Boston, Massachus
- Page 167 and 168:
LETTERSInfluenza A(H5N6)Virus Reass
- Page 169 and 170:
LETTERSsystem (8 kb-span paired-end
- Page 171 and 172:
LETTERS3. Van Hong N, Amambua-Ngwa
- Page 173 and 174:
LETTERSTable. Prevalence of Bartone
- Page 175 and 176:
LETTERSavian influenza A(H5N1) viru
- Page 177 and 178:
LETTERSprovinces and a total of 200
- Page 179 and 180:
LETTERS7. Manian FA. Bloodstream in
- Page 181 and 182:
LETTERSforward projections. N Engl
- Page 183 and 184:
LETTERS3. Guindon S, Gascuel OA. Si
- Page 185 and 186:
BOOKS AND MEDIAin the port cities o
- Page 187 and 188:
ABOUT THE COVERNorth was not intere
- Page 189 and 190:
Earning CME CreditTo obtain credit,
- Page 191:
Emerging Infectious Diseases is a p