Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
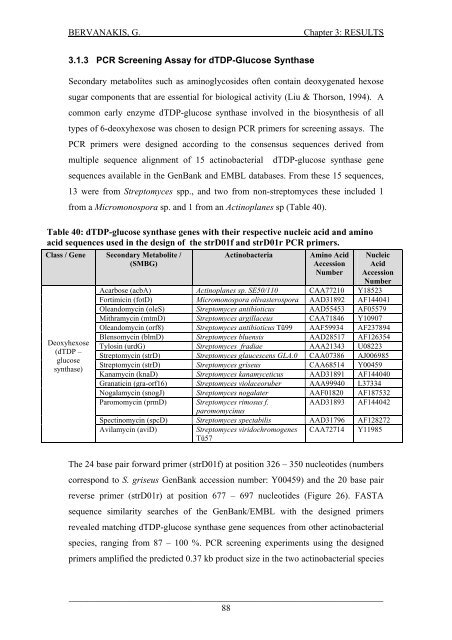
BERVANAKIS, G.Chapter 3: RESULTS3.1.3 PCR Screen<strong>in</strong>g Assay for dTDP-Glucose SynthaseSecondary metabolites such as am<strong>in</strong>oglycosides <strong>of</strong>ten conta<strong>in</strong> deoxygenated hexosesugar components that are essential for biological activity (Liu & Thorson, 1994). Acommon early enzyme dTDP-glucose synthase <strong>in</strong>volved <strong>in</strong> the biosynthesis <strong>of</strong> alltypes <strong>of</strong> 6-deoxyhexose was chosen to design PCR primers for screen<strong>in</strong>g assays. ThePCR primers were designed accord<strong>in</strong>g to the consensus sequences derived frommultiple sequence alignment <strong>of</strong> 15 act<strong>in</strong>obacterial dTDP-glucose synthase genesequences available <strong>in</strong> the GenBank <strong>and</strong> EMBL databases. From these 15 sequences,13 were from Streptomyces spp., <strong>and</strong> two from non-streptomyces these <strong>in</strong>cluded 1from a Micromonospora sp. <strong>and</strong> 1 from an Act<strong>in</strong>oplanes sp (Table 40).Table 40: dTDP-glucose synthase genes with their respective nucleic acid <strong>and</strong> am<strong>in</strong>oacid sequences used <strong>in</strong> the design <strong>of</strong> the strD01f <strong>and</strong> strD01r PCR primers.Class / Gene Secondary Metabolite /(SMBG)Deoxyhexose(dTDP –glucosesynthase)Act<strong>in</strong>obacteriaAm<strong>in</strong>o AcidAccessionNumberNucleicAcidAccessionNumberAcarbose (acbA) Act<strong>in</strong>oplanes sp. SE50/110 CAA77210 Y18523Fortimic<strong>in</strong> (fotD) Micromonospora olivasterospora AAD31892 AF144041Ole<strong>and</strong>omyc<strong>in</strong> (oleS) Streptomyces antibioticus AAD55453 AF05579Mithramyc<strong>in</strong> (mtmD) Streptomyces argillaceus CAA71846 Y10907Ole<strong>and</strong>omyc<strong>in</strong> (orf8) Streptomyces antibioticus Tü99 AAF59934 AF237894Blensomyc<strong>in</strong> (blmD) Streptomyces bluensis AAD28517 AF126354Tylos<strong>in</strong> (urdG) Streptomyces fradiae AAA21343 U08223Streptomyc<strong>in</strong> (strD) Streptomyces glaucescens GLA.0 CAA07386 AJ006985Streptomyc<strong>in</strong> (strD) Streptomyces griseus CAA68514 Y00459Kanamyc<strong>in</strong> (knaD) Streptomyces kanamyceticus AAD31891 AF144040Granatic<strong>in</strong> (gra-orf16) Streptomyces violaceoruber AAA99940 L37334Nogalamyc<strong>in</strong> (snogJ) Streptomyces nogalater AAF01820 AF187532Paromomyc<strong>in</strong> (prmD) Streptomyces rimosus f.AAD31893 AF144042paromomyc<strong>in</strong>usSpect<strong>in</strong>omyc<strong>in</strong> (spcD) Streptomyces spectabilis AAD31796 AF128272Avilamyc<strong>in</strong> (aviD)Streptomyces viridochromogenes CAA72714 Y11985Tü57The 24 base pair forward primer (strD01f) at position 326 – 350 nucleotides (numberscorrespond to S. griseus GenBank accession number: Y00459) <strong>and</strong> the 20 base pairreverse primer (strD01r) at position 677 – 697 nucleotides (Figure 26). FASTAsequence similarity searches <strong>of</strong> the GenBank/EMBL with the designed primersrevealed match<strong>in</strong>g dTDP-glucose synthase gene sequences from other act<strong>in</strong>obacterialspecies, rang<strong>in</strong>g from 87 – 100 %. PCR screen<strong>in</strong>g experiments us<strong>in</strong>g the designedprimers amplified the predicted 0.37 kb product size <strong>in</strong> the two act<strong>in</strong>obacterial species_____________________________________________________________________88