Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
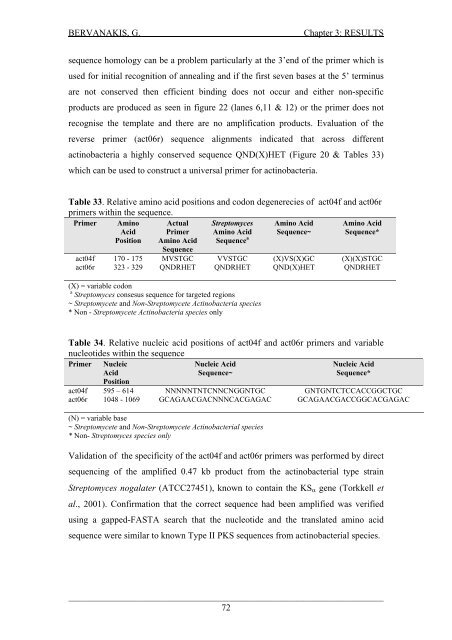
BERVANAKIS, G.Chapter 3: RESULTSsequence homology can be a problem particularly at the 3’end <strong>of</strong> the primer which isused for <strong>in</strong>itial recognition <strong>of</strong> anneal<strong>in</strong>g <strong>and</strong> if the first seven bases at the 5’ term<strong>in</strong>usare not conserved then efficient b<strong>in</strong>d<strong>in</strong>g does not occur <strong>and</strong> either non-specificproducts are produced as seen <strong>in</strong> figure 22 (lanes 6,11 & 12) or the primer does notrecognise the template <strong>and</strong> there are no amplification products. Evaluation <strong>of</strong> thereverse primer (act06r) sequence alignments <strong>in</strong>dicated that across differentact<strong>in</strong>obacteria a highly conserved sequence QND(X)HET (Figure 20 & Tables 33)which can be used to construct a universal primer for act<strong>in</strong>obacteria.Table 33. Relative am<strong>in</strong>o acid positions <strong>and</strong> codon degenerecies <strong>of</strong> act04f <strong>and</strong> act06rprimers with<strong>in</strong> the sequence.Primer Am<strong>in</strong>oAcidActualPrimerStreptomycesAm<strong>in</strong>o AcidAm<strong>in</strong>o AcidSequence~Am<strong>in</strong>o AcidSequence*Position Am<strong>in</strong>o AcidSequenceSequence aact04f 170 - 175 MVSTGC VVSTGC (X)VS(X)GC (X)(X)STGCact06r 323 - 329 QNDRHET QNDRHET QND(X)HET QNDRHET(X) = variable codona Streptomyces consesus sequence for targeted regions~ Streptomycete <strong>and</strong> Non-Streptomycete Act<strong>in</strong>obacteria species* Non - Streptomycete Act<strong>in</strong>obacteria species onlyTable 34. Relative nucleic acid positions <strong>of</strong> act04f <strong>and</strong> act06r primers <strong>and</strong> variablenucleotides with<strong>in</strong> the sequencePrimer NucleicAcidNucleic AcidSequence~Nucleic AcidSequence*Positionact04f 595 – 614 NNNNNTNTCNNCNGGNTGC GNTGNTCTCCACCGGCTGCact06r 1048 - 1069 GCAGAACGACNNNCACGAGAC GCAGAACGACCGGCACGAGAC(N) = variable base~ Streptomycete <strong>and</strong> Non-Streptomycete Act<strong>in</strong>obacterial species* Non- Streptomyces species onlyValidation <strong>of</strong> the specificity <strong>of</strong> the act04f <strong>and</strong> act06r primers was performed by directsequenc<strong>in</strong>g <strong>of</strong> the amplified 0.47 kb product from the act<strong>in</strong>obacterial type stra<strong>in</strong>Streptomyces nogalater (ATCC27451), known to conta<strong>in</strong> the KS α gene (Torkkell etal., 2001). Confirmation that the correct sequence had been amplified was verifiedus<strong>in</strong>g a gapped-FASTA search that the nucleotide <strong>and</strong> the translated am<strong>in</strong>o acidsequence were similar to known Type II PKS sequences from act<strong>in</strong>obacterial species.___________________________________________________________________________________72