Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
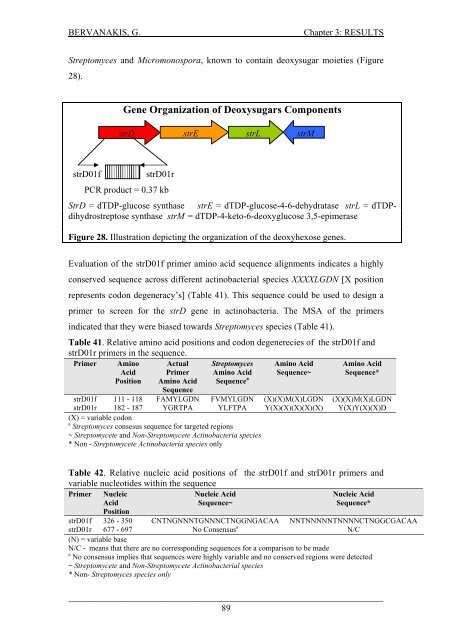
BERVANAKIS, G.Chapter 3: RESULTSStreptomyces <strong>and</strong> Micromonospora, known to conta<strong>in</strong> deoxysugar moieties (Figure28).Gene Organization <strong>of</strong> Deoxysugars ComponentsstrD strE strL strMstrD01fstrD01rPCR product = 0.37 kbStrD = dTDP-glucose synthase strE = dTDP-glucose-4-6-dehydratase strL = dTDPdihydrostreptosesynthase strM = dTDP-4-keto-6-deoxyglucose 3,5-epimeraseFigure 28. Illustration depict<strong>in</strong>g the organization <strong>of</strong> the deoxyhexose genes.Evaluation <strong>of</strong> the strD01f primer am<strong>in</strong>o acid sequence alignments <strong>in</strong>dicates a highlyconserved sequence across different act<strong>in</strong>obacterial species XXXXLGDN [X positionrepresents codon degeneracy’s] (Table 41). This sequence could be used to design aprimer to screen for the strD gene <strong>in</strong> act<strong>in</strong>obacteria. The MSA <strong>of</strong> the primers<strong>in</strong>dicated that they were biased towards Streptomyces species (Table 41).Table 41. Relative am<strong>in</strong>o acid positions <strong>and</strong> codon degenerecies <strong>of</strong> the strD01f <strong>and</strong>strD01r primers <strong>in</strong> the sequence.Primer Am<strong>in</strong>oAcidActualPrimerStreptomycesAm<strong>in</strong>o AcidAm<strong>in</strong>o AcidSequence~Am<strong>in</strong>o AcidSequence*Position Am<strong>in</strong>o AcidSequenceSequence astrD01f 111 - 118 FAMYLGDN FVMYLGDN (X)(X)M(X)LGDN (X)(X)M(X)LGDNstrD01r 182 - 187 YGRTPA YLFTPA Y(X)(X)(X)(X)(X) Y(X)Y(X)(X)D(X) = variable codona Streptomyces consesus sequence for targeted regions~ Streptomycete <strong>and</strong> Non-Streptomycete Act<strong>in</strong>obacteria species* Non - Streptomycete Act<strong>in</strong>obacteria species onlyTable 42. Relative nucleic acid positions <strong>of</strong> the strD01f <strong>and</strong> strD01r primers <strong>and</strong>variable nucleotides with<strong>in</strong> the sequencePrimer NucleicAcidNucleic AcidSequence~Nucleic AcidSequence*PositionstrD01f 326 - 350 CNTNGNNNTGNNNCTNGGNGACAA NNTNNNNNTNNNNCTNGGCGACAAstrD01r 677 - 697 No Consensus a N/C(N) = variable baseN/C - means that there are no corresspond<strong>in</strong>g sequences for a comparison to be madea No consensus implies that sequences were highly variable <strong>and</strong> no conserved regions were detected~ Streptomycete <strong>and</strong> Non-Streptomycete Act<strong>in</strong>obacterial species* Non- Streptomyces species only_____________________________________________________________________89