Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
Detection and Expression of Biosynthetic Genes in Actinobacteria ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
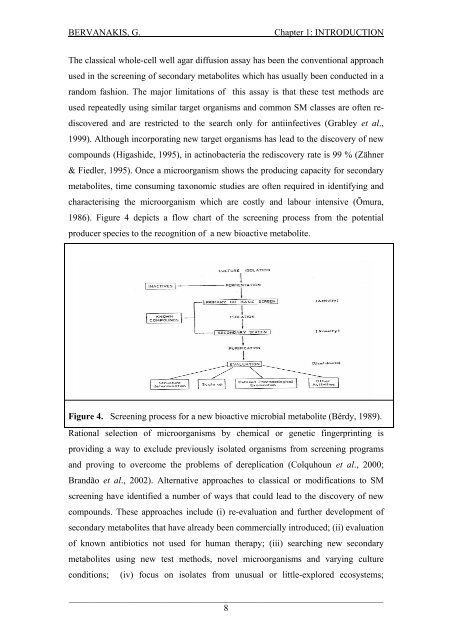
BERVANAKIS, G.Chapter 1: INTRODUCTIONThe classical whole-cell well agar diffusion assay has been the conventional approachused <strong>in</strong> the screen<strong>in</strong>g <strong>of</strong> secondary metabolites which has usually been conducted <strong>in</strong> ar<strong>and</strong>om fashion. The major limitations <strong>of</strong> this assay is that these test methods areused repeatedly us<strong>in</strong>g similar target organisms <strong>and</strong> common SM classes are <strong>of</strong>ten rediscovered<strong>and</strong> are restricted to the search only for anti<strong>in</strong>fectives (Grabley et al.,1999). Although <strong>in</strong>corporat<strong>in</strong>g new target organisms has lead to the discovery <strong>of</strong> newcompounds (Higashide, 1995), <strong>in</strong> act<strong>in</strong>obacteria the rediscovery rate is 99 % (Zähner& Fiedler, 1995). Once a microorganism shows the produc<strong>in</strong>g capacity for secondarymetabolites, time consum<strong>in</strong>g taxonomic studies are <strong>of</strong>ten required <strong>in</strong> identify<strong>in</strong>g <strong>and</strong>characteris<strong>in</strong>g the microorganism which are costly <strong>and</strong> labour <strong>in</strong>tensive (Ōmura,1986). Figure 4 depicts a flow chart <strong>of</strong> the screen<strong>in</strong>g process from the potentialproducer species to the recognition <strong>of</strong> a new bioactive metabolite.Figure 4. Screen<strong>in</strong>g process for a new bioactive microbial metabolite (Bérdy, 1989).Rational selection <strong>of</strong> microorganisms by chemical or genetic f<strong>in</strong>gerpr<strong>in</strong>t<strong>in</strong>g isprovid<strong>in</strong>g a way to exclude previously isolated organisms from screen<strong>in</strong>g programs<strong>and</strong> prov<strong>in</strong>g to overcome the problems <strong>of</strong> dereplication (Colquhoun et al., 2000;Br<strong>and</strong>ão et al., 2002). Alternative approaches to classical or modifications to SMscreen<strong>in</strong>g have identified a number <strong>of</strong> ways that could lead to the discovery <strong>of</strong> newcompounds. These approaches <strong>in</strong>clude (i) re-evaluation <strong>and</strong> further development <strong>of</strong>secondary metabolites that have already been commercially <strong>in</strong>troduced; (ii) evaluation<strong>of</strong> known antibiotics not used for human therapy; (iii) search<strong>in</strong>g new secondarymetabolites us<strong>in</strong>g new test methods, novel microorganisms <strong>and</strong> vary<strong>in</strong>g cultureconditions; (iv) focus on isolates from unusual or little-explored ecosystems;_____________________________________________________________________8