December 2012 Number 1 - Utah Native Plant Society
December 2012 Number 1 - Utah Native Plant Society
December 2012 Number 1 - Utah Native Plant Society
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Calochortiana <strong>December</strong> <strong>2012</strong> <strong>Number</strong> 1<br />
MATERIALS AND METHODS<br />
Sample Collection<br />
Phacelia argillacea leaf tissue samples were collected<br />
from Tucker in the 2006 and 2008 field seasons<br />
and from Railroad in the 2006, 2007, and 2008 field<br />
seasons. A single basal leaf was removed nondestructively<br />
from each individual. The 2004 Tucker samples<br />
represent half-sibling progeny from wild-collected seeds<br />
of 15 maternal individuals (a total of 53 plants) in the<br />
2004 field season. These individuals were grown for<br />
seed production. Phacelia crenulata samples represented<br />
individuals greenhouse-grown from seeds of a<br />
bulk wild collection. Phacelia argylensis and some P.<br />
glandulosa samples were collected from Brigham<br />
Young University Herbarium; specimens were annotated<br />
as sampled for this study. The remaining P. glandulosa<br />
samples represent bulk population samples from<br />
two closely adjacent populations collected by Frank<br />
Smith in 2007 (Figure 1).<br />
DNA Extraction and AFLP Analysis<br />
Fresh leaf tissue samples for DNA extraction were<br />
dried over silica gel, lypophilized, or frozen at -80C immediately<br />
after collection. DNA was extracted from<br />
tissue samples using a Qiagen <strong>Plant</strong> Mini Kit (QIAGEN,<br />
Inc., Valencia, CA) with minor modifications in the protocol<br />
to achieve a higher concentration of DNA.<br />
AFLP analysis was carried out following Vos et al<br />
(1995) with minor modifications. The enzymes EcoRI<br />
and MseI were used for DNA digestion. Each plant<br />
sample was fingerprinted with six primer combinations.<br />
The primer extensions used were EcoAA/MseA,<br />
EcoAA/MseG, EcoAA/MseT, EcoAC/MseA, EcoAC/<br />
MseG, and EcoAC/MseT. Fragment separation and detection<br />
was carried out on a LI-COR 4300 DNA Analysis<br />
System (LI-COR Biosciences, Lincoln, NE) on a<br />
6.5% polyacrylamide gel. Only unambiguous bands<br />
(50 – 350 bp) were scored for presence or absence.<br />
Bands that were monomorphic among all samples were<br />
discarded from analysis of polymorphic bands. Principal<br />
components analysis was performed on the complete<br />
data set, on data from the three close congeners alone,<br />
on data from P. argillacea alone, and on data from the<br />
Tucker half-sib families alone. We used SAS software<br />
(SAS Institute, Cary, NC) for the analysis.<br />
RESULTS<br />
AFLP analysis produced a total of 535 reliably reproducible<br />
bands, 124 of which were polymorphic. Seventy-five<br />
of these bands were polymorphic only between<br />
P. crenulata and the other Phacelia species (P. glandulosa,<br />
P. argylensis, and P. argillacea; Table 1). This<br />
clearly demonstrated that the P. glandulosa group is<br />
strongly genetically differentiated from P. crenulata, the<br />
putative distant congener in the study. The three close<br />
congeners were much more genetically similar. Phacelia<br />
argillacea exhibited nine bands that were polymorphic<br />
with P. argylensis, seven polymorphic bands<br />
within P. glandulosa from herbarium material, and<br />
seven polymorphic bands within P. glandulosa collected<br />
by Frank Smith in western Colorado. An unexpected<br />
result was that the Smith collections were even more<br />
differentiated from other P. glandulosa than was P. argillacea,<br />
with 15 bands polymorphic between the two<br />
groups. In contrast, P. argylensis was closely similar to<br />
the herbarium-collected P. glandulosa group, with only<br />
3 polymorphic bands. Within P. argillacea, we observed<br />
a total of 30 polymorphic bands, however no polymorphic<br />
bands were found between the Tucker and<br />
Railroad populations, suggesting low genetic differentiation.<br />
When we analyzed data from all four Phacelia species<br />
included in the study, the first principal component<br />
represented 79% of the total variation and clearly separated<br />
P. crenulata form the other three species, reflect-<br />
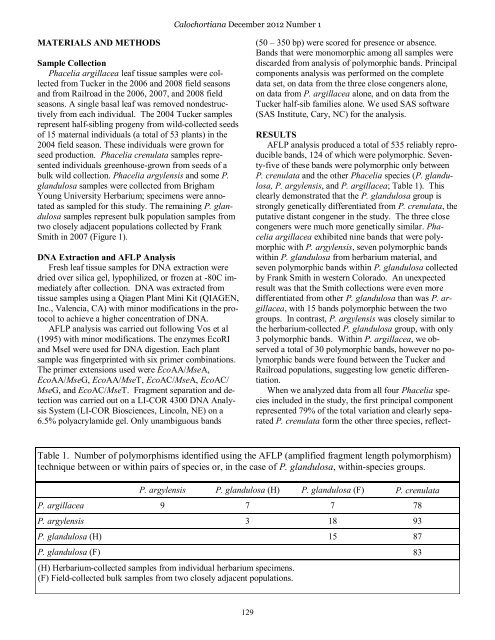
Table 1. <strong>Number</strong> of polymorphisms identified using the AFLP (amplified fragment length polymorphism)<br />
technique between or within pairs of species or, in the case of P. glandulosa, within-species groups.<br />
P. argylensis P. glandulosa (H) P. glandulosa (F) P. crenulata<br />
P. argillacea 9 7 7 78<br />
P. argylensis 3 18 93<br />
P. glandulosa (H) 15 87<br />
P. glandulosa (F) 83<br />
(H) Herbarium-collected samples from individual herbarium specimens.<br />
(F) Field-collected bulk samples from two closely adjacent populations.<br />
129