research activities in 2007 - CSEM
research activities in 2007 - CSEM
research activities in 2007 - CSEM
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Simultaneous Detection of Four Antibiotic Families <strong>in</strong> Milk for Customer Safety<br />
G. Voir<strong>in</strong>, R. Ischer, S. Pasche<br />
A biosensor for the detection of four antibiotic families has been realized and tested with reference milk samples <strong>in</strong> collaboration with European<br />
partners. The simultaneous detection and identification of antibiotics <strong>in</strong> contam<strong>in</strong>ated milk has been demonstrated.<br />
In the dairy <strong>in</strong>dustry, it is important to avoid contam<strong>in</strong>ation of<br />
milk with antibiotics which could modify the fermentation<br />
process of dairy products such as cheese or yogurt, or<br />
possibly cause allergic reactions <strong>in</strong> consumers (Figure 1). In<br />
the frame of the European project GoodFood [1] , a biosensor<br />
system for the detection of several antibiotics has been<br />
developed <strong>in</strong> collaboration with other European partners.<br />
<strong>CSEM</strong> has focused on a biosensor detection system based on<br />
the <strong>in</strong>terrogation of the resonance wavelength of a waveguide<br />
grat<strong>in</strong>g coupler (WIOS Wavelength Interrogated Optical<br />
Sens<strong>in</strong>g). The relevant antibiotics were identified dur<strong>in</strong>g a<br />
survey made <strong>in</strong> 30 countries around the world. Four antibiotic<br />
families were selected: sulphonamides, fluoroqu<strong>in</strong>olones,<br />
beta-lactams and tetracycl<strong>in</strong>es.<br />
Figure 1: Milk cha<strong>in</strong> from production to consumer, screen<strong>in</strong>g test<br />
must be performed as early as possible<br />
The detection system is based on the change of the refractive<br />
<strong>in</strong>dex of the sens<strong>in</strong>g surface due to the b<strong>in</strong>d<strong>in</strong>g of molecules.<br />
The refractive <strong>in</strong>dex change <strong>in</strong>duces a shift <strong>in</strong> the resonance<br />
wavelength of the waveguide grat<strong>in</strong>g coupler which is<br />
<strong>in</strong>terrogated us<strong>in</strong>g a periodically swept tunable laser. This<br />
detection method is sensitive to refractive <strong>in</strong>dex changes on<br />
the order of 10-6 , or the equivalent of several pg/mm2 of<br />
molecules b<strong>in</strong>d<strong>in</strong>g to the sensor surface.<br />
A competitive immunoassay format was chosen for the<br />
detection of the antibiotics. A specific sens<strong>in</strong>g surface was<br />
obta<strong>in</strong>ed by us<strong>in</strong>g recognition molecules such as antibodies<br />
and receptors, developed by GoodFood partners. Specific<br />
antibodies for the sulphonamide and fluoroqu<strong>in</strong>olone antibiotic<br />
families were obta<strong>in</strong>ed by immunization of rabbits, and are<br />
specific for a whole family of antibiotics (a family is a group of<br />
molecules with similar chemical structures). Receptor<br />
molecules specific for the beta-lactams and tetracycl<strong>in</strong>e<br />
antibiotic families were specially eng<strong>in</strong>eered to present a high<br />
aff<strong>in</strong>ity for a group of molecules. For each family, a test<br />
protocol was implemented on the biosensor platform and the<br />
cross reactions with the other reagents and antibiotics were<br />
tested. The goal was to obta<strong>in</strong> a biosensor platform with<br />
different sens<strong>in</strong>g regions each specific to one antibiotic family,<br />
allow<strong>in</strong>g detection <strong>in</strong> the different regions simultaneously with<br />
a unique milk sample. Us<strong>in</strong>g optical detection allows<br />
measurements on eight separate pads simultaneously.<br />
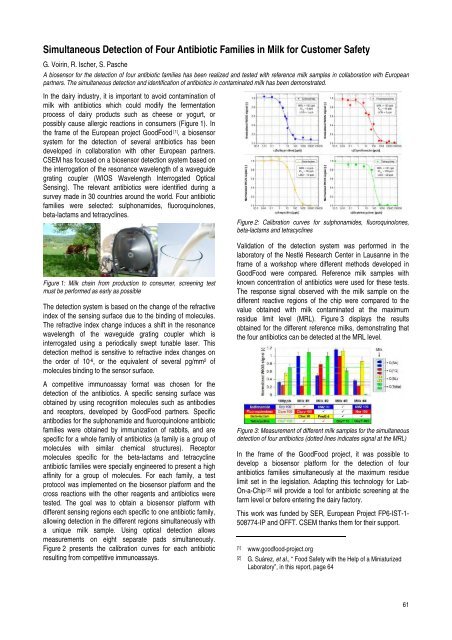
Figure 2 presents the calibration curves for each antibiotic<br />
result<strong>in</strong>g from competitive immunoassays.<br />
Figure 2: Calibration curves for sulphonamides, fluoroqu<strong>in</strong>olones,<br />
beta-lactams and tetracycl<strong>in</strong>es<br />
Validation of the detection system was performed <strong>in</strong> the<br />
laboratory of the Nestlé Research Center <strong>in</strong> Lausanne <strong>in</strong> the<br />
frame of a workshop where different methods developed <strong>in</strong><br />
GoodFood were compared. Reference milk samples with<br />
known concentration of antibiotics were used for these tests.<br />
The response signal observed with the milk sample on the<br />
different reactive regions of the chip were compared to the<br />
value obta<strong>in</strong>ed with milk contam<strong>in</strong>ated at the maximum<br />
residue limit level (MRL). Figure 3 displays the results<br />
obta<strong>in</strong>ed for the different reference milks, demonstrat<strong>in</strong>g that<br />
the four antibiotics can be detected at the MRL level.<br />
Figure 3: Measurement of different milk samples for the simultaneous<br />
detection of four antibiotics (dotted l<strong>in</strong>es <strong>in</strong>dicates signal at the MRL)<br />
In the frame of the GoodFood project, it was possible to<br />
develop a biosensor platform for the detection of four<br />
antibiotics families simultaneously at the maximum residue<br />
limit set <strong>in</strong> the legislation. Adapt<strong>in</strong>g this technology for Lab-<br />
On-a-Chip [2] will provide a tool for antibiotic screen<strong>in</strong>g at the<br />
farm level or before enter<strong>in</strong>g the dairy factory.<br />
This work was funded by SER, European Project FP6-IST-1-<br />
508774-IP and OFFT. <strong>CSEM</strong> thanks them for their support.<br />
[1] www.goodfood-project.org<br />
[2] G. Suárez, et al., “ Food Safety with the Help of a M<strong>in</strong>iaturized<br />
Laboratory”, <strong>in</strong> this report, page 64<br />
61