Research Group Heussler (Malaria I) - Bernhard-Nocht-Institut für ...
Research Group Heussler (Malaria I) - Bernhard-Nocht-Institut für ...
Research Group Heussler (Malaria I) - Bernhard-Nocht-Institut für ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
39<br />
Parasitology Section<br />
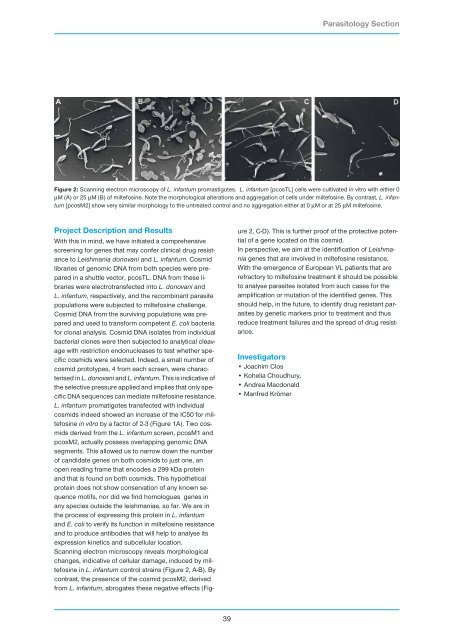
Figure 2: Scanning electron microscopy of L. infantum promastigotes. L. infantum [pcosTL] cells were cultivated in vitro with either 0<br />
µM (A) or 25 µM (B) of miltefosine. Note the morphological alterations and aggregation of cells under miltefosine. By contrast, L. infantum<br />
[pcosM2] show very similar morphology to the untreated control and no aggregation either at 0 µM or at 25 µM miltefosine.<br />
Project Description and Results<br />
With this in mind, we have initiated a comprehensive<br />
screening for genes that may confer clinical drug resistance<br />
to Leishmania donovani and L. infantum. Cosmid<br />
libraries of genomic DNA from both species were prepared<br />
in a shuttle vector, pcosTL. DNA from these libraries<br />
were electrotransfected into L. donovani and<br />
L. infantum, respectively, and the recombinant parasite<br />
populations were subjected to miltefosine challenge.<br />
Cosmid DNA from the surviving populations was prepared<br />
and used to transform competent E. coli bacteria<br />
for clonal analysis. Cosmid DNA isolates from individual<br />
bacterial clones were then subjected to analytical cleavage<br />
with restriction endonucleases to test whether specific<br />
cosmids were selected. Indeed, a small number of<br />
cosmid prototypes, 4 from each screen, were characterised<br />
in L. donovani and L. infantum. This is indicative of<br />
the selective pressure applied and implies that only specific<br />
DNA sequences can mediate miltefosine resistance.<br />
L. infantum promatigotes transfected with individual<br />
cosmids indeed showed an increase of the IC50 for miltefosine<br />
in vitro by a factor of 2-3 (Figure 1A). Two cosmids<br />
derived from the L. infantum screen, pcosM1 and<br />
pcosM2, actually possess overlapping genomic DNA<br />
segments. This allowed us to narrow down the number<br />
of candidate genes on both cosmids to just one, an<br />
open reading frame that encodes a 299 kDa protein<br />
and that is found on both cosmids. This hypothetical<br />
protein does not show conservation of any known sequence<br />
motifs, nor did we find homologues genes in<br />
any species outside the leishmaniae, so far. We are in<br />
the process of expressing this protein in L. infantum<br />
and E. coli to verify its function in miltefosine resistance<br />
and to produce antibodies that will help to analyse its<br />
expression kinetics and subcellular location.<br />
Scanning electron microscopy reveals morphological<br />
changes, indicative of cellular damage, induced by miltefosine<br />
in L. infantum control strains (Figure 2, A-B). By<br />
contrast, the presence of the cosmid pcosM2, derived<br />
from L. infantum, abrogates these negative effects (Fig-<br />
ure 2, C-D). This is further proof of the protective potential<br />
of a gene located on this cosmid.<br />
In perspective, we aim at the identification of Leishmania<br />
genes that are involved in miltefosine resistance.<br />
With the emergence of European VL patients that are<br />
refractory to miltefosine treatment it should be possible<br />
to analyse parasites isolated from such cases for the<br />
amplification or mutation of the identified genes. This<br />
should help, in the future, to identify drug resistant parasites<br />
by genetic markers prior to treatment and thus<br />
reduce treatment failures and the spread of drug resistance.<br />
Investigators<br />
• Joachim Clos<br />
• Kohelia Choudhury,<br />
• Andrea Macdonald<br />
• Manfred Krömer