GTMB 7 - Gene Therapy & Molecular Biology
GTMB 7 - Gene Therapy & Molecular Biology
GTMB 7 - Gene Therapy & Molecular Biology
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
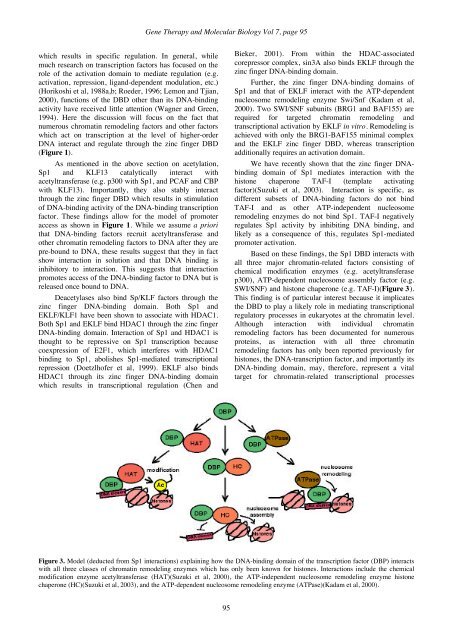
<strong>Gene</strong> <strong>Therapy</strong> and <strong>Molecular</strong> <strong>Biology</strong> Vol 7, page 95which results in specific regulation. In general, whilemuch research on transcription factors has focused on therole of the activation domain to mediate regulation (e.g.activation, repression, ligand-dependent modulation, etc.)(Horikoshi et al, 1988a,b; Roeder, 1996; Lemon and Tjian,2000), functions of the DBD other than its DNA-bindingactivity have received little attention (Wagner and Green,1994). Here the discussion will focus on the fact thatnumerous chromatin remodeling factors and other factorswhich act on transcription at the level of higher-orderDNA interact and regulate through the zinc finger DBD(Figure 1).As mentioned in the above section on acetylation,Sp1 and KLF13 catalytically interact withacetyltransferase (e.g. p300 with Sp1, and PCAF and CBPwith KLF13). Importantly, they also stably interactthrough the zinc finger DBD which results in stimulationof DNA-binding activity of the DNA-binding transcriptionfactor. These findings allow for the model of promoteraccess as shown in Figure 1. While we assume a priorithat DNA-binding factors recruit acetyltransferase andother chromatin remodeling factors to DNA after they arepre-bound to DNA, these results suggest that they in factshow interaction in solution and that DNA binding isinhibitory to interaction. This suggests that interactionpromotes access of the DNA-binding factor to DNA but isreleased once bound to DNA.Deacetylases also bind Sp/KLF factors through thezinc finger DNA-binding domain. Both Sp1 andEKLF/KLF1 have been shown to associate with HDAC1.Both Sp1 and EKLF bind HDAC1 through the zinc fingerDNA-binding domain. Interaction of Sp1 and HDAC1 isthought to be repressive on Sp1 transcription becausecoexpression of E2F1, which interferes with HDAC1binding to Sp1, abolishes Sp1-mediated transcriptionalrepression (Doetzlhofer et al, 1999). EKLF also bindsHDAC1 through its zinc finger DNA-binding domainwhich results in transcriptional regulation (Chen andBieker, 2001). From within the HDAC-associatedcorepressor complex, sin3A also binds EKLF through thezinc finger DNA-binding domain.Further, the zinc finger DNA-binding domains ofSp1 and that of EKLF interact with the ATP-dependentnucleosome remodeling enzyme Swi/Snf (Kadam et al,2000). Two SWI/SNF subunits (BRG1 and BAF155) arerequired for targeted chromatin remodeling andtranscriptional activation by EKLF in vitro. Remodeling isachieved with only the BRG1-BAF155 minimal complexand the EKLF zinc finger DBD, whereas transcriptionadditionally requires an activation domain.We have recently shown that the zinc finger DNAbindingdomain of Sp1 mediates interaction with thehistone chaperone TAF-I (template activatingfactor)(Suzuki et al, 2003). Interaction is specific, asdifferent subsets of DNA-binding factors do not bindTAF-I and as other ATP-independent nucleosomeremodeling enzymes do not bind Sp1. TAF-I negativelyregulates Sp1 activity by inhibiting DNA binding, andlikely as a consequence of this, regulates Sp1-mediatedpromoter activation.Based on these findings, the Sp1 DBD interacts withall three major chromatin-related factors consisting ofchemical modification enzymes (e.g. acetyltransferasep300), ATP-dependent nucleosome assembly factor (e.g.SWI/SNF) and histone chaperone (e.g. TAF-I)(Figure 3).This finding is of particular interest because it implicatesthe DBD to play a likely role in mediating transcriptionalregulatory processes in eukaryotes at the chromatin level.Although interaction with individual chromatinremodeling factors has been documented for numerousproteins, as interaction with all three chromatinremodeling factors has only been reported previously forhistones, the DNA-transcription factor, and importantly itsDNA-binding domain, may, therefore, represent a vitaltarget for chromatin-related transcriptional processesFigure 3. Model (deducted from Sp1 interactions) explaining how the DNA-binding domain of the transcription factor (DBP) interactswith all three classes of chromatin remodeling enzymes which has only been known for histones. Interactions include the chemicalmodification enzyme acetyltransferase (HAT)(Suzuki et al, 2000), the ATP-independent nucleosome remodeling enzyme histonechaperone (HC)(Suzuki et al, 2003), and the ATP-dependent nucleosome remodeling enzyme (ATPase)(Kadam et al, 2000).95