GTMB 7 - Gene Therapy & Molecular Biology
GTMB 7 - Gene Therapy & Molecular Biology
GTMB 7 - Gene Therapy & Molecular Biology
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
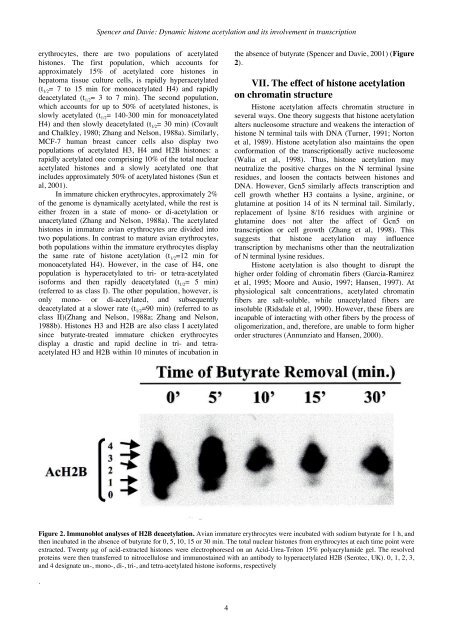
Spencer and Davie: Dynamic histone acetylation and its involvement in transcriptionerythrocytes, there are two populations of acetylatedhistones. The first population, which accounts forapproximately 15% of acetylated core histones inhepatoma tissue culture cells, is rapidly hyperacetylated(t 1/2 = 7 to 15 min for monoacetylated H4) and rapidlydeacetylated (t 1/2 = 3 to 7 min). The second population,which accounts for up to 50% of acetylated histones, isslowly acetylated (t 1/2 = 140-300 min for monoacetylatedH4) and then slowly deacetylated (t 1/2 = 30 min) (Covaultand Chalkley, 1980; Zhang and Nelson, 1988a). Similarly,MCF-7 human breast cancer cells also display twopopulations of acetylated H3, H4 and H2B histones: arapidly acetylated one comprising 10% of the total nuclearacetylated histones and a slowly acetylated one thatincludes approximately 50% of acetylated histones (Sun etal, 2001).In immature chicken erythrocytes, approximately 2%of the genome is dynamically acetylated, while the rest iseither frozen in a state of mono- or di-acetylation orunacetylated (Zhang and Nelson, 1988a). The acetylatedhistones in immature avian erythrocytes are divided intotwo populations. In contrast to mature avian erythrocytes,both populations within the immature erythrocytes displaythe same rate of histone acetylation (t 1/2 =12 min formonoacetylated H4). However, in the case of H4, onepopulation is hyperacetylated to tri- or tetra-acetylatedisoforms and then rapidly deacetylated (t 1/2 = 5 min)(referred to as class I). The other population, however, isonly mono- or di-acetylated, and subsequentlydeacetylated at a slower rate (t 1/2 =90 min) (referred to asclass II)(Zhang and Nelson, 1988a; Zhang and Nelson,1988b). Histones H3 and H2B are also class I acetylatedsince butyrate-treated immature chicken erythrocytesdisplay a drastic and rapid decline in tri- and tetraacetylatedH3 and H2B within 10 minutes of incubation inthe absence of butyrate (Spencer and Davie, 2001) (Figure2).VII. The effect of histone acetylationon chromatin structureHistone acetylation affects chromatin structure inseveral ways. One theory suggests that histone acetylationalters nucleosome structure and weakens the interaction ofhistone N terminal tails with DNA (Turner, 1991; Nortonet al, 1989). Histone acetylation also maintains the openconformation of the transcriptionally active nucleosome(Walia et al, 1998). Thus, histone acetylation mayneutralize the positive charges on the N terminal lysineresidues, and loosen the contacts between histones andDNA. However, Gcn5 similarly affects transcription andcell growth whether H3 contains a lysine, arginine, orglutamine at position 14 of its N terminal tail. Similarly,replacement of lysine 8/16 residues with arginine orglutamine does not alter the affect of Gcn5 ontranscription or cell growth (Zhang et al, 1998). Thissuggests that histone acetylation may influencetranscription by mechanisms other than the neutralizationof N terminal lysine residues.Histone acetylation is also thought to disrupt thehigher order folding of chromatin fibers (Garcia-Ramirezet al, 1995; Moore and Ausio, 1997; Hansen, 1997). Atphysiological salt concentrations, acetylated chromatinfibers are salt-soluble, while unacetylated fibers areinsoluble (Ridsdale et al, 1990). However, these fibers areincapable of interacting with other fibers by the process ofoligomerization, and, therefore, are unable to form higherorder structures (Annunziato and Hansen, 2000).Figure 2. Immunoblot analyses of H2B deacetylation. Avian immature erythrocytes were incubated with sodium butyrate for 1 h, andthen incubated in the absence of butyrate for 0, 5, 10, 15 or 30 min. The total nuclear histones from erythrocytes at each time point wereextracted. Twenty µg of acid-extracted histones were electrophoresed on an Acid-Urea-Triton 15% polyacrylamide gel. The resolvedproteins were then transferred to nitrocellulose and immunostained with an antibody to hyperacetylated H2B (Serotec, UK). 0, 1, 2, 3,and 4 designate un-, mono-, di-, tri-, and tetra-acetylated histone isoforms, respectively.4