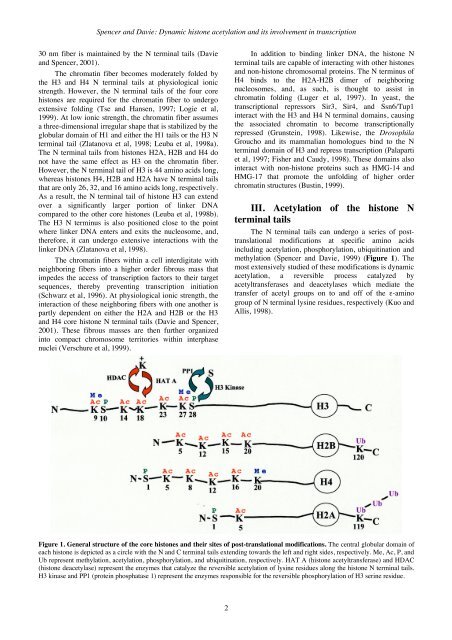
Spencer and Davie: Dynamic histone acetylation and its involvement in transcription30 nm fiber is maintained by the N terminal tails (Davieand Spencer, 2001).The chromatin fiber becomes moderately folded bythe H3 and H4 N terminal tails at physiological ionicstrength. However, the N terminal tails of the four corehistones are required for the chromatin fiber to undergoextensive folding (Tse and Hansen, 1997; Logie et al,1999). At low ionic strength, the chromatin fiber assumesa three-dimensional irregular shape that is stabilized by theglobular domain of H1 and either the H1 tails or the H3 Nterminal tail (Zlatanova et al, 1998; Leuba et al, 1998a).The N terminal tails from histones H2A, H2B and H4 donot have the same effect as H3 on the chromatin fiber.However, the N terminal tail of H3 is 44 amino acids long,whereas histones H4, H2B and H2A have N terminal tailsthat are only 26, 32, and 16 amino acids long, respectively.As a result, the N terminal tail of histone H3 can extendover a significantly larger portion of linker DNAcompared to the other core histones (Leuba et al, 1998b).The H3 N terminus is also positioned close to the pointwhere linker DNA enters and exits the nucleosome, and,therefore, it can undergo extensive interactions with thelinker DNA (Zlatanova et al, 1998).The chromatin fibers within a cell interdigitate withneighboring fibers into a higher order fibrous mass thatimpedes the access of transcription factors to their targetsequences, thereby preventing transcription initiation(Schwarz et al, 1996). At physiological ionic strength, theinteraction of these neighboring fibers with one another ispartly dependent on either the H2A and H2B or the H3and H4 core histone N terminal tails (Davie and Spencer,2001). These fibrous masses are then further organizedinto compact chromosome territories within interphasenuclei (Verschure et al, 1999).In addition to binding linker DNA, the histone Nterminal tails are capable of interacting with other histonesand non-histone chromosomal proteins. The N terminus ofH4 binds to the H2A-H2B dimer of neighboringnucleosomes, and, as such, is thought to assist inchromatin folding (Luger et al, 1997). In yeast, thetranscriptional repressors Sir3, Sir4, and Ssn6/Tup1interact with the H3 and H4 N terminal domains, causingthe associated chromatin to become transcriptionallyrepressed (Grunstein, 1998). Likewise, the DrosophilaGroucho and its mammalian homologues bind to the Nterminal domain of H3 and repress transcription (Palapartiet al, 1997; Fisher and Caudy, 1998). These domains alsointeract with non-histone proteins such as HMG-14 andHMG-17 that promote the unfolding of higher orderchromatin structures (Bustin, 1999).III. Acetylation of the histone Nterminal tailsThe N terminal tails can undergo a series of posttranslationalmodifications at specific amino acidsincluding acetylation, phosphorylation, ubiquitination andmethylation (Spencer and Davie, 1999) (Figure 1). Themost extensively studied of these modifications is dynamicacetylation, a reversible process catalyzed byacetyltransferases and deacetylases which mediate thetransfer of acetyl groups on to and off of the ε-aminogroup of N terminal lysine residues, respectively (Kuo andAllis, 1998).Figure 1. <strong>Gene</strong>ral structure of the core histones and their sites of post-translational modifications. The central globular domain ofeach histone is depicted as a circle with the N and C terminal tails extending towards the left and right sides, respectively. Me, Ac, P, andUb represent methylation, acetylation, phosphorylation, and ubiquitination, respectively. HAT A (histone acetyltransferase) and HDAC(histone deacetylase) represent the enzymes that catalyze the reversible acetylation of lysine residues along the histone N terminal tails.H3 kinase and PP1 (protein phosphatase 1) represent the enzymes responsible for the reversible phosphorylation of H3 serine residue.2
<strong>Gene</strong> <strong>Therapy</strong> and <strong>Molecular</strong> <strong>Biology</strong> Vol 7, page 3This modification typically occurs on up to fivelysine residues along the H3 and H4 N terminal tails, fourresidues along H2B, and one residue along H2A (Davieand Spencer, 1999). Whether a histone is hypo- orhyperacetylated depends on the net activities ofneighboring histone acetyltransferases and deacetylases.IV. Histone acetyltransferasesThe following is only a brief summary of the histoneacetyltransferases identified to date. For a more detaileddescription of histone acetyltransferases and theirsubstrates, please refer to the following reviews (Sternerand Berger, 2000; Davie and Spencer, 2001; Marmorsteinand Roth, 2001; Bertos et al, 2001). Numeroustranscription co-activators including yGcn5, P/CAF,CBP/p300, Esa1, NuA4, and ACTR/SRC-1 have beenidentified as having intrinsic histone acetyltransferaseactivity (Sterner and Berger, 2000; Davie and Spencer,2001; Klochendler-Yeivin and Yaniv, 2001; Marmorsteinand Roth, 2001). In addition, the DNA-bindingtransactivator ATF-2, the general transcription factorsTAFII250 and Nut1, and the elongation factor Elp3 arehistone acetyltransferases (Marmorstein and Roth, 2001).Histone acetyltransferases generally exist in largecomplexes (Spencer and Davie, 1999). Each histoneacetyltransferase has a different target substrate, and thespecificity for this substrate depends on the proteinsassociated with the histone acetyltransferase (Grant et al,1999). For example, the free full-length form of yeastGcn5 preferentially acetylates H3 in vitro and H3 and H4in vivo (Zhang et al, 1998; Sterner and Berger, 2000;Davie and Spencer, 2001). However, the acetylatingefficiency of yeast Gcn5 for nucleosomal histonesincreases when assembled into high molecular weight,multi-protein complexes referred to as SAGA (Spt-Ada-Gcn5-acetyltransferase) and Ada (Grant et al, 1999). Inaddition, the pattern of histone acetylation for Gcn5assembled into the SAGA complex is distinct from thatexhibited by Gcn5 when assembled into Ada (Grant et al,1999). Similarly, the histone substrate specificity ofindividual human PCAF and yeast Esa1 acetyltransferasesbecomes altered when these enzymes are assembled intomulti-protein complexes (Davie and Spencer, 2001). Thephosphorylation of CBP by ERK1 enhances the activity ofthis acetyltransferase, suggesting that the function ofhistone acetyltransferases may be regulated byphosphorylation events (Liu et al, 1999).V. Histone deacetylasesAs many as 10 histone deacetylases have beenidentified to date (Bertos et al, 2001). Refer to thefollowing reviews (Sterner and Berger, 2000; Bertos et al,2001; Davie and Spencer, 2001; Marmorstein and Roth,2001) for a more detailed description of histonedeacetylases. These deacetylases are divided into 3 classesdefined by their size and sequence homologies to yeastdeacetylases. The class I histone deacetylases areapproximately 400-500 amino acids in length and includeHDACs 1,2,3 and 8. These class I members are nucleartranscriptional co-repressors with homology to the yeastRpd3 deacetylase. The class II histone deacetylases arelarger proteins of approximately 1000 amino acids withstructural homology to yeast Hda1 and include HDACs4,5,6,7,9 and 10 (Davie and Moniwa, 2000; Bertos et al,2001; Guardiola and Yao, 2002). Class III histonedeacetylases are encoded by genes similar to the yeastsilent information regulator (Sir 2) gene (Afshar andMurnane, 1999; Frye, 1999). These deacetylases aredependent on NAD+ and ADP-ribosylase activity (Frye,2000; Imai et al, 2000; Landry et al, 2000).Class I deacetylases are ubiquitously expressed,while class II deacetylases are tissue-, cell-anddifferentiation-specific (Davie and Moniwa, 2000). Bothclasses of deacetylases can deacetylate the four corehistones, however, each deacetylase has a site preference(Davie and Spencer, 2001). Similar to histoneacetyltransferases, the yeast Rpd3 and Hda1 deacetylasesexist in distinct multi-protein complexes, suggesting thatclass I and II deacetylases have distinct biologicalfunctions. Furthermore, the components of thesecomplexes influence the substrate specificity of theseenzymes (Davie and Moniwa, 2000). For example, the freeform of avian HDAC1 preferentially deacetylates free butnot nucleosomal H3. When assembled into a multi-proteincomplex, this deacetylase preferentially deacetylates freeH2B and histones assembled into a nucleosome (Sun et al,1999).Class I deacetylases reside in the nucleus (Davie andMoniwa, 2000). However, the sub-cellular distribution ofclass II deacetylases is not as straight forward. HDACs 4and 5 shuttle between the cytoplasm and the nucleus(Bertos et al, 2001). HDAC7 is predominantly nuclear butbinds to the membrane-associated endothelin receptor Aand most likely functions in the cytoplasm (Lee et al,2001). HDAC6 is strictly cytoplasmic, and HDAC9appears to be both nuclear and cytoplasmic (Zhou et al,2001). HDACs 4,5, and 7 are transcriptional co-repressorsthat interact with MEF2 transcription factors, as well asthe co-repressors N-CoR, BCoR, and CtBP (Bertos et al,2001; Guardiola and Yao, 2002). Similarly, HDAC9interacts with MEF-2 and represses MEF-2-mediatedtranscription (Zhou et al, 2001). HDAC10 resides in thenucleus and the cytoplasm (Guardiola and Yao, 2002). Inthe nucleus, this deacetylase functions as a transcriptionalrepressor when tethered to a promoter (Guardiola and Yao,2002). Interestingly, HDAC6 can interact with ubiquitin.As well, the mammalian homologue of UFD3, a yeastprotein involved in protein ubiquitination, is part of thecytoplasmic mammalian HDAC6 complex (Seigneurin-Berny et al, 2001).VI. The dynamics of histoneacetylationStudies of histone acetylation dynamics indicate thatboth acetylation and deacetylation occur at more than onerate (Covault and Chalkley, 1980; Zhang and Nelson,1988a). In human fibroblasts and mature avian3
- Page 7 and 8: Instructions to authors:Gene Therap
- Page 9: Please submit an electronic version
- Page 12: 103-111 ResearchArticle113-133 Revi
- Page 19 and 20: Gene Therapy and Molecular Biology
- Page 21 and 22: Gene Therapy and Molecular Biology
- Page 23 and 24: Gene Therapy and Molecular Biology
- Page 25 and 26: Gene Therapy and Molecular Biology
- Page 27 and 28: Gene Therapy and Molecular Biology
- Page 29 and 30: Gene Therapy and Molecular Biology
- Page 31 and 32: Gene Therapy and Molecular Biology
- Page 33 and 34: Gene Therapy and Molecular Biology
- Page 35 and 36: Gene Therapy and Molecular Biology
- Page 37 and 38: Gene Therapy and Molecular Biology
- Page 39 and 40: Gene Therapy and Molecular Biology
- Page 41 and 42: Gene Therapy and Molecular Biology
- Page 43 and 44: Gene Therapy and Molecular Biology
- Page 45 and 46: Gene Therapy and Molecular Biology
- Page 47 and 48: Gene Therapy and Molecular Biology
- Page 49 and 50: Gene Therapy and Molecular Biology
- Page 51 and 52: Gene Therapy and Molecular Biology
- Page 53 and 54: Gene Therapy and Molecular Biology
- Page 55 and 56: Gene Therapy and Molecular Biology
- Page 57 and 58: Gene Therapy and Molecular Biology
- Page 59 and 60: Gene Therapy and Molecular Biology
- Page 61 and 62: Gene Therapy and Molecular Biology
- Page 63 and 64:
Gene Therapy and Molecular Biology
- Page 65 and 66:
Gene Therapy and Molecular Biology
- Page 67 and 68:
Gene Therapy and Molecular Biology
- Page 69 and 70:
Gene Therapy and Molecular Biology
- Page 71 and 72:
Gene Therapy and Molecular Biology
- Page 73 and 74:
Gene Therapy and Molecular Biology
- Page 75 and 76:
Gene Therapy and Molecular Biology
- Page 77:
Gene Therapy and Molecular Biology
- Page 80 and 81:
Epperly et al: Late injection of Mn
- Page 82 and 83:
Epperly et al: Late injection of Mn
- Page 84 and 85:
Goldberg-Cohen et al: Regulation of
- Page 86 and 87:
Goldberg-Cohen et al: Regulation of
- Page 88 and 89:
Goldberg-Cohen et al: Regulation of
- Page 90 and 91:
Gascón-Irún et al: Gene therapy a
- Page 92 and 93:
Gascón-Irún et al: Gene therapy a
- Page 94 and 95:
Gascón-Irún et al: Gene therapy a
- Page 96 and 97:
Gascón-Irún et al: Gene therapy a
- Page 98 and 99:
Gascón-Irún et al: Gene therapy a
- Page 100 and 101:
Gascón-Irún et al: Gene therapy a
- Page 102 and 103:
Gascón-Irún et al: Gene therapy a
- Page 104 and 105:
Gascón-Irún et al: Gene therapy a
- Page 106 and 107:
Suzuki et al: Regulation of the Sp/
- Page 108 and 109:
Suzuki et al: Regulation of the Sp/
- Page 110 and 111:
Suzuki et al: Regulation of the Sp/
- Page 112 and 113:
Suzuki et al: Regulation of the Sp/
- Page 114 and 115:
Li et al: MET amplification in live
- Page 116 and 117:
Li et al: MET amplification in live
- Page 118 and 119:
Chavakis et al: Leukocyte adhesion
- Page 120 and 121:
Chavakis et al: Leukocyte adhesion
- Page 122 and 123:
Chavakis et al: Leukocyte adhesion
- Page 124 and 125:
Chavakis et al: Leukocyte adhesion
- Page 126 and 127:
Chavakis et al: Leukocyte adhesion
- Page 128 and 129:
Sanlioglu et al: Adenovirus mediate
- Page 130 and 131:
Sanlioglu et al: Adenovirus mediate
- Page 132 and 133:
Sanlioglu et al: Adenovirus mediate
- Page 134 and 135:
Sanlioglu et al: Adenovirus mediate
- Page 136 and 137:
Sanlioglu et al: Adenovirus mediate
- Page 138 and 139:
Sanlioglu et al: Adenovirus mediate
- Page 140 and 141:
Sanlioglu et al: Adenovirus mediate
- Page 142 and 143:
Sanlioglu et al: Adenovirus mediate
- Page 144 and 145:
Sanlioglu et al: Adenovirus mediate
- Page 146 and 147:
Sanlioglu et al: Adenovirus mediate
- Page 148 and 149:
Sanlioglu et al: Adenovirus mediate
- Page 150 and 151:
George et al: Gene therapy for vasc
- Page 152 and 153:
George et al: Gene therapy for vasc
- Page 154 and 155:
George et al: Gene therapy for vasc
- Page 156 and 157:
George et al: Gene therapy for vasc
- Page 158 and 159:
George et al: Gene therapy for vasc
- Page 160 and 161:
George et al: Gene therapy for vasc
- Page 162 and 163:
George et al: Gene therapy for vasc
- Page 164 and 165:
George et al: Gene therapy for vasc
- Page 166 and 167:
George et al: Gene therapy for vasc
- Page 168 and 169:
Zhang et al: Angiogenic Gene Therap
- Page 170 and 171:
Zhang et al: Angiogenic Gene Therap
- Page 172 and 173:
Zhang et al: Angiogenic Gene Therap
- Page 174 and 175:
Zhang et al: Angiogenic Gene Therap
- Page 176 and 177:
Zhang et al: Angiogenic Gene Therap
- Page 178 and 179:
Zhang et al: Angiogenic Gene Therap
- Page 180 and 181:
Zhang et al: Angiogenic Gene Therap
- Page 182 and 183:
Xu et al: G-CSF receptor-mediated S
- Page 184 and 185:
Xu et al: G-CSF receptor-mediated S
- Page 186 and 187:
Xu et al: G-CSF receptor-mediated S
- Page 188 and 189:
Burek et al: Calcium induced cell d
- Page 190 and 191:
Burek et al: Calcium induced cell d
- Page 192 and 193:
Burek et al: Calcium induced cell d
- Page 194 and 195:
Burek et al: Calcium induced cell d
- Page 196 and 197:
David et al: Current status and fut
- Page 198 and 199:
David et al: Current status and fut
- Page 200 and 201:
David et al: Current status and fut
- Page 202 and 203:
David et al: Current status and fut
- Page 204 and 205:
David et al: Current status and fut
- Page 206 and 207:
David et al: Current status and fut
- Page 208 and 209:
David et al: Current status and fut
- Page 210 and 211:
David et al: Current status and fut
- Page 212 and 213:
David et al: Current status and fut
- Page 214 and 215:
David et al: Current status and fut
- Page 216 and 217:
David et al: Current status and fut
- Page 218 and 219:
David et al: Current status and fut
- Page 220 and 221:
David et al: Current status and fut
- Page 222 and 223:
David et al: Current status and fut
- Page 224 and 225:
David et al: Current status and fut
- Page 226 and 227:
Stoll et al: The role of EBV and ge
- Page 228 and 229:
Stoll et al: The role of EBV and ge
- Page 230 and 231:
Stoll et al: The role of EBV and ge
- Page 232 and 233:
Stoll et al: The role of EBV and ge
- Page 234 and 235:
Stoll et al: The role of EBV and ge
- Page 236 and 237:
Maruyama et al: Kidney-targeted pla
- Page 238 and 239:
Maruyama et al: Kidney-targeted pla
- Page 240 and 241:
Maruyama et al: Kidney-targeted pla
- Page 242 and 243:
Maruyama et al: Kidney-targeted pla
- Page 244 and 245:
Kren et al: Hepatocyte-targeted del
- Page 246 and 247:
Kren et al: Hepatocyte-targeted del
- Page 248 and 249:
Kren et al: Hepatocyte-targeted del
- Page 250 and 251:
Kren et al: Hepatocyte-targeted del
- Page 252 and 253:
Kren et al: Hepatocyte-targeted del
- Page 254 and 255:
Zeng: PRL-3 as a target for cancer
- Page 256 and 257:
Zeng: PRL-3 as a target for cancer
- Page 258 and 259:
Zeng: PRL-3 as a target for cancer
- Page 260 and 261:
Latchman: Protective effect of heat
- Page 262 and 263:
Latchman: Protective effect of heat
- Page 264 and 265:
Latchman: Protective effect of heat
- Page 266 and 267:
Latchman: Protective effect of heat
- Page 268 and 269:
Latchman: Protective effect of heat
- Page 270 and 271:
Cai et al: Lung cancer gene therapy
- Page 272 and 273:
Cai et al: Lung cancer gene therapy
- Page 274 and 275:
Cai et al: Lung cancer gene therapy
- Page 276 and 277:
Cai et al: Lung cancer gene therapy
- Page 278 and 279:
Cai et al: Lung cancer gene therapy
- Page 280:
Cai et al: Lung cancer gene therapy
- Page 283 and 284:
Gene Therapy and Molecular Biology
- Page 285 and 286:
Gene Therapy and Molecular Biology
- Page 287 and 288:
Gene Therapy and Molecular Biology
- Page 289 and 290:
Gene Therapy and Molecular Biology
- Page 291 and 292:
Gene Therapy and Molecular Biology
- Page 293 and 294:
Gene Therapy and Molecular Biology
- Page 295 and 296:
Gene Therapy and Molecular Biology
- Page 297 and 298:
Gene Therapy and Molecular Biology
- Page 299 and 300:
Gene Therapy and Molecular Biology
- Page 301 and 302:
Gene Therapy and Molecular Biology
- Page 303 and 304:
Gene Therapy and Molecular Biology
- Page 305 and 306:
Gene Therapy and Molecular Biology
- Page 307 and 308:
Gene Therapy and Molecular Biology
- Page 309:
Gene Therapy and Molecular Biology