NUI Galway – UL Alliance First Annual ENGINEERING AND - ARAN ...
NUI Galway – UL Alliance First Annual ENGINEERING AND - ARAN ...
NUI Galway – UL Alliance First Annual ENGINEERING AND - ARAN ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Improving discovery in Life Sciences and Health Care with Semantic Web<br />
Technologies and Linked Data<br />
Helena F. Deus and Jonas S. Almeida<br />
Digital Enterprise Research Institute, National University of Ireland, <strong>Galway</strong><br />
Email: helena.deus@deri.org<br />
Abstract<br />
One of the most enticing outcomes of biological<br />
exploration for the quantitative-minded researcher is to<br />
identify the “mathematics of biology” through the study<br />
of the patterns and interrelatedness of biological<br />
entities. To move forward in that direction, many pieces<br />
need to be set in place, from the availability of<br />
biological data in forms that can be computationally<br />
manipulated to the automated discovery of patterns that<br />
would derive from integration of data in many and<br />
diverse endpoints. A computational system where<br />
researchers could securely deposit any type of data and<br />
have it immediately analyzed, traversed, annotated and<br />
merged with data deposited elsewhere is a dream not<br />
yet achieved but one which could revolutionize<br />
scientific discovery and, ultimately, help cure disease.<br />
1. Introduction<br />
“Science is organized knowledge”, said Immanuel<br />
Kant. Science in the Information Age has indeed been<br />
characterized by increasingly complex approaches to the<br />
organization and sharing of scientific knowledge to<br />
improve its discovery. A critical bottleneck in applying<br />
knowledge engineering to inform and improve<br />
biomedical discovery is the ability to align experimental<br />
data acquisition with knowledge representation models.<br />
Recent technological advances in genomics, proteomics<br />
and other ‘omics’ sciences, have flooded microbiology<br />
labs with unprecedented amounts of experimental data<br />
and were at the heart of a paradigm shift for researchers<br />
interested in its computational representation and<br />
analysis.<br />
2. Knowledge Continuum for Life Sciences<br />
The deluge of data in biology marked the beginning<br />
of the “industrialization of data production in Life<br />
Sciences beyond a craft-based cottage industry” [1].<br />
The successful exploit of this new wave of data<br />
acquisition technologies created the need for multiple,<br />
overlapping, sub-disciplines of biology to cope with the<br />
increasing complexity of biological results. However,<br />
this became an obstacle to discovery because the<br />
answers to biological problems often span multiple<br />
domain boundaries and rely on the existence of<br />
knowledge continuums. Starting to address these<br />
challenges requires agreement on optimal strategies for<br />
publishing experimental biomedical data in<br />
interoperable formats even before they are made<br />
available to the community through peer-reviewed<br />
publication.<br />
129<br />
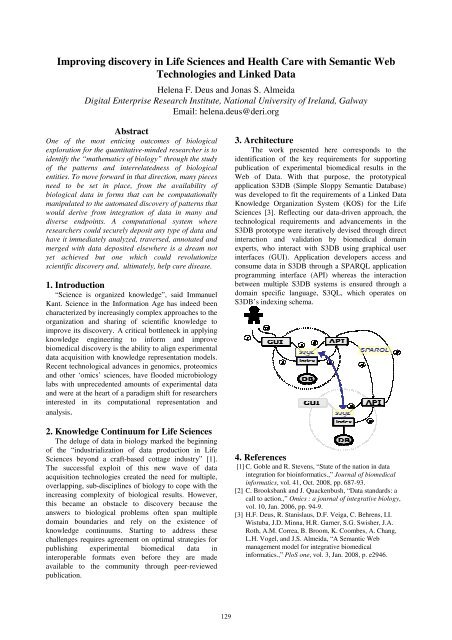
3. Architecture<br />
The work presented here corresponds to the<br />
identification of the key requirements for supporting<br />
publication of experimental biomedical results in the<br />
Web of Data. With that purpose, the prototypical<br />
application S3DB (Simple Sloppy Semantic Database)<br />
was developed to fit the requirements of a Linked Data<br />
Knowledge Organization System (KOS) for the Life<br />
Sciences [3]. Reflecting our data-driven approach, the<br />
technological requirements and advancements in the<br />
S3DB prototype were iteratively devised through direct<br />
interaction and validation by biomedical domain<br />
experts, who interact with S3DB using graphical user<br />
interfaces (GUI). Application developers access and<br />
consume data in S3DB through a SPARQL application<br />
programming interface (API) whereas the interaction<br />
between multiple S3DB systems is ensured through a<br />
domain specific language, S3QL, which operates on<br />
S3DB’s indexing schema.<br />
4. References<br />
[1] C. Goble and R. Stevens, “State of the nation in data<br />
integration for bioinformatics.,” Journal of biomedical<br />
informatics, vol. 41, Oct. 2008, pp. 687-93.<br />
[2] C. Brooksbank and J. Quackenbush, “Data standards: a<br />
call to action.,” Omics : a journal of integrative biology,<br />
vol. 10, Jan. 2006, pp. 94-9.<br />
[3] H.F. Deus, R. Stanislaus, D.F. Veiga, C. Behrens, I.I.<br />
Wistuba, J.D. Minna, H.R. Garner, S.G. Swisher, J.A.<br />
Roth, A.M. Correa, B. Broom, K. Coombes, A. Chang,<br />
L.H. Vogel, and J.S. Almeida, “A Semantic Web<br />
management model for integrative biomedical<br />
informatics.,” PloS one, vol. 3, Jan. 2008, p. e2946.