- Page 7 and 8:
ContentsPrefaceChapter IChapter IIC
- Page 9 and 10:
PrefaceThe 6-carbon lactone known a
- Page 11 and 12:
Prefaceixbalance of antioxidant and
- Page 13 and 14:
Prefacexiprovided or is contradicto
- Page 15 and 16:
PrefacexiiiChapter IX - Background:
- Page 17 and 18:
Prefacexvdetoxification defence sys
- Page 19:
Prefacexviiprogressed faster than i
- Page 22 and 23:
2Kelsey H. Fisher-Wellman and Richa
- Page 24 and 25:
4Kelsey H. Fisher-Wellman and Richa
- Page 26 and 27:
6Kelsey H. Fisher-Wellman and Richa
- Page 28 and 29:
8Kelsey H. Fisher-Wellman and Richa
- Page 30 and 31:
10Kelsey H. Fisher-Wellman and Rich
- Page 32 and 33:
12Kelsey H. Fisher-Wellman and Rich
- Page 34 and 35:
14Kelsey H. Fisher-Wellman and Rich
- Page 36 and 37:
16Kelsey H. Fisher-Wellman and Rich
- Page 38 and 39:
18Kelsey H. Fisher-Wellman and Rich
- Page 40 and 41:
20Kelsey H. Fisher-Wellman and Rich
- Page 42 and 43:
22Kelsey H. Fisher-Wellman and Rich
- Page 44 and 45:
24Kelsey H. Fisher-Wellman and Rich
- Page 46 and 47:
26Kelsey H. Fisher-Wellman and Rich
- Page 48 and 49:
28Kelsey H. Fisher-Wellman and Rich
- Page 50 and 51:
30Kelsey H. Fisher-Wellman and Rich
- Page 52 and 53:
32Kelsey H. Fisher-Wellman and Rich
- Page 54 and 55:
34Kelsey H. Fisher-Wellman and Rich
- Page 56 and 57:
36Kelsey H. Fisher-Wellman and Rich
- Page 58 and 59:
38Kelsey H. Fisher-Wellman and Rich
- Page 60 and 61:
40Kelsey H. Fisher-Wellman and Rich
- Page 62 and 63:
42Kelsey H. Fisher-Wellman and Rich
- Page 65 and 66:
In: Handbook of Vitamin C Research
- Page 67 and 68:
Human Specific Vitamin C Metabolism
- Page 69 and 70:
Human Specific Vitamin C Metabolism
- Page 71 and 72:
Human Specific Vitamin C Metabolism
- Page 73 and 74:
Human Specific Vitamin C Metabolism
- Page 75 and 76:
Human Specific Vitamin C Metabolism
- Page 77 and 78:
Human Specific Vitamin C Metabolism
- Page 79 and 80:
Human Specific Vitamin C Metabolism
- Page 81 and 82:
mgmgmgHuman Specific Vitamin C Meta
- Page 83 and 84:
Human Specific Vitamin C Metabolism
- Page 85 and 86:
Human Specific Vitamin C Metabolism
- Page 87 and 88:
Human Specific Vitamin C Metabolism
- Page 89 and 90:
Human Specific Vitamin C Metabolism
- Page 91 and 92:
Human Specific Vitamin C Metabolism
- Page 93 and 94:
Human Specific Vitamin C Metabolism
- Page 95 and 96:
Human Specific Vitamin C Metabolism
- Page 97 and 98:
Human Specific Vitamin C Metabolism
- Page 99 and 100:
Human Specific Vitamin C Metabolism
- Page 101 and 102:
Human Specific Vitamin C Metabolism
- Page 103 and 104:
Human Specific Vitamin C Metabolism
- Page 105:
Human Specific Vitamin C Metabolism
- Page 108 and 109:
88Ana I. Haza, Almudena García and
- Page 110 and 111:
90Figure 1.Ana I. Haza, Almudena Ga
- Page 112 and 113:
92Ana I. Haza, Almudena García and
- Page 114 and 115:
94Figure 3.Ana I. Haza, Almudena Ga
- Page 116 and 117:
96Ana I. Haza, Almudena García and
- Page 118 and 119:
98Ana I. Haza, Almudena García and
- Page 120 and 121:
100Ana I. Haza, Almudena García an
- Page 122 and 123:
102Ana I. Haza, Almudena García an
- Page 124 and 125:
104Ana I. Haza, Almudena García an
- Page 126 and 127:
106Ana I. Haza, Almudena García an
- Page 128 and 129:
108Ana I. Haza, Almudena García an
- Page 130 and 131:
110Ana I. Haza, Almudena García an
- Page 132 and 133:
112Ana I. Haza, Almudena García an
- Page 134 and 135:
114Ana I. Haza, Almudena García an
- Page 136 and 137:
116Ana I. Haza, Almudena García an
- Page 138 and 139:
118Ana I. Haza, Almudena García an
- Page 140 and 141:
120Ana I. Haza, Almudena García an
- Page 142 and 143:
122Ana I. Haza, Almudena García an
- Page 144 and 145:
124Ana I. Haza, Almudena García an
- Page 147 and 148:
In: Handbook of Vitamin C Research:
- Page 149 and 150:
Vitamin C: Dietary Requirements, Di
- Page 151 and 152:
Oxidative DamageVitamin C: Dietary
- Page 153 and 154:
Vitamin C: Dietary Requirements, Di
- Page 155 and 156:
Vitamin C: Dietary Requirements, Di
- Page 157 and 158:
Vitamin C: Dietary Requirements, Di
- Page 159 and 160:
Vitamin C: Dietary Requirements, Di
- Page 161 and 162:
Vitamin C: Dietary Requirements, Di
- Page 163 and 164:
Vitamin C: Dietary Requirements, Di
- Page 165 and 166:
Vitamin C: Dietary Requirements, Di
- Page 167 and 168:
Vitamin C: Dietary Requirements, Di
- Page 169 and 170:
Vitamin C: Dietary Requirements, Di
- Page 171 and 172:
Vitamin C: Dietary Requirements, Di
- Page 173 and 174:
In: Handbook of Vitamin C Research:
- Page 175 and 176:
Pro-Oxidant vs. Antioxidant Effects
- Page 177 and 178:
Pro-Oxidant vs. Antioxidant Effects
- Page 179 and 180:
Pro-Oxidant vs. Antioxidant Effects
- Page 181 and 182:
Pro-Oxidant vs. Antioxidant Effects
- Page 183 and 184:
Patient. B.H. (38), atopic eczema.V
- Page 185 and 186:
Pro-Oxidant vs. Antioxidant Effects
- Page 187 and 188: Pro-Oxidant vs. Antioxidant Effects
- Page 189 and 190: Pro-Oxidant vs. Antioxidant Effects
- Page 191 and 192: Pro-Oxidant vs. Antioxidant Effects
- Page 193 and 194: Pro-Oxidant vs. Antioxidant Effects
- Page 195 and 196: Pro-Oxidant vs. Antioxidant Effects
- Page 197 and 198: Pro-Oxidant vs. Antioxidant Effects
- Page 199 and 200: Pro-Oxidant vs. Antioxidant Effects
- Page 201 and 202: Pro-Oxidant vs. Antioxidant Effects
- Page 203: Pro-Oxidant vs. Antioxidant Effects
- Page 206 and 207: 186Magdalena Stevanović and Dragan
- Page 208 and 209: 188Magdalena Stevanović and Dragan
- Page 210 and 211: 190Magdalena Stevanović and Dragan
- Page 212 and 213: 192Magdalena Stevanović and Dragan
- Page 214 and 215: 194Magdalena Stevanović and Dragan
- Page 216 and 217: 196Magdalena Stevanović and Dragan
- Page 218 and 219: 198Magdalena Stevanović and Dragan
- Page 220 and 221: 200Magdalena Stevanović and Dragan
- Page 222 and 223: 202Magdalena Stevanović and Dragan
- Page 224 and 225: 204Magdalena Stevanović and Dragan
- Page 226 and 227: 206Magdalena Stevanović and Dragan
- Page 228 and 229: 208Magdalena Stevanović and Dragan
- Page 230 and 231: 210Magdalena Stevanović and Dragan
- Page 233 and 234: In: Handbook of Vitamin C Research
- Page 235 and 236: Molecular Bases of the Cellular Han
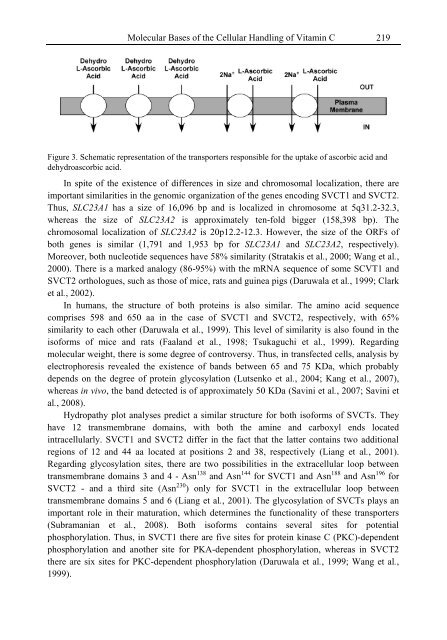
- Page 237: Molecular Bases of the Cellular Han
- Page 241 and 242: Molecular Bases of the Cellular Han
- Page 243 and 244: Molecular Bases of the Cellular Han
- Page 245 and 246: Molecular Bases of the Cellular Han
- Page 247 and 248: Molecular Bases of the Cellular Han
- Page 249 and 250: Molecular Bases of the Cellular Han
- Page 251 and 252: Molecular Bases of the Cellular Han
- Page 253 and 254: Molecular Bases of the Cellular Han
- Page 255 and 256: Molecular Bases of the Cellular Han
- Page 257 and 258: In: Handbook of Vitamin C Research
- Page 259 and 260: Vitamin C: Daily Requirements, Diet
- Page 261 and 262: Vitamin C: Daily Requirements, Diet
- Page 263 and 264: Vitamin C: Daily Requirements, Diet
- Page 265 and 266: Vitamin C: Daily Requirements, Diet
- Page 267 and 268: Vitamin C: Daily Requirements, Diet
- Page 269 and 270: Vitamin C: Daily Requirements, Diet
- Page 271 and 272: Vitamin C: Daily Requirements, Diet
- Page 273 and 274: Vitamin C: Daily Requirements, Diet
- Page 275 and 276: Vitamin C: Daily Requirements, Diet
- Page 277 and 278: Vitamin C: Daily Requirements, Diet
- Page 279: Vitamin C: Daily Requirements, Diet
- Page 282 and 283: 262Alan M. Preston, Luis Vázquez Q
- Page 284 and 285: 264Alan M. Preston, Luis Vázquez Q
- Page 286 and 287: 266Alan M. Preston, Luis Vázquez Q
- Page 288 and 289:
Percent of Study Group Selecting Di
- Page 290 and 291:
270Alan M. Preston, Luis Vázquez Q
- Page 292 and 293:
272Alan M. Preston, Luis Vázquez Q
- Page 294 and 295:
274Alan M. Preston, Luis Vázquez Q
- Page 296 and 297:
276Alan M. Preston, Luis Vázquez Q
- Page 298 and 299:
278Alan M. Preston, Luis Vázquez Q
- Page 301 and 302:
In: Handbook of Vitamin C Research
- Page 303 and 304:
The Role of Vitamin C in Human Repr
- Page 305 and 306:
The Role of Vitamin C in Human Repr
- Page 307 and 308:
The Role of Vitamin C in Human Repr
- Page 309 and 310:
The Role of Vitamin C in Human Repr
- Page 311 and 312:
The Role of Vitamin C in Human Repr
- Page 313 and 314:
The Role of Vitamin C in Human Repr
- Page 315 and 316:
The Role of Vitamin C in Human Repr
- Page 317:
The Role of Vitamin C in Human Repr
- Page 320 and 321:
300Mustafa NazıroğluKeywords: Vit
- Page 322 and 323:
302Mustafa NazıroğluNO is synthes
- Page 324 and 325:
304Mustafa Nazıroğlu‗recycling
- Page 326 and 327:
306Mustafa Nazıroğluagents, such
- Page 328 and 329:
308Mustafa NazıroğluFuture Direct
- Page 330 and 331:
310Mustafa Nazıroğlu[21] Basu, TK
- Page 332 and 333:
312Mustafa Nazıroğlu[59] Cengiz M
- Page 334 and 335:
314Samar Al Sayegh Petkovšek and B
- Page 336 and 337:
316Samar Al Sayegh Petkovšek and B
- Page 338 and 339:
318Samar Al Sayegh Petkovšek and B
- Page 340 and 341:
(mgg -1 )320Samar Al Sayegh Petkov
- Page 342 and 343:
322Samar Al Sayegh Petkovšek and B
- Page 344 and 345:
324Samar Al Sayegh Petkovšek and B
- Page 346 and 347:
326Samar Al Sayegh Petkovšek and B
- Page 348 and 349:
328Samar Al Sayegh Petkovšek and B
- Page 350 and 351:
330Antonio J. López-Farré, José
- Page 352 and 353:
332Antonio J. López-Farré, José
- Page 354 and 355:
334Antonio J. López-Farré, José
- Page 356 and 357:
336Antonio J. López-Farré, José
- Page 358 and 359:
338Antonio J. López-Farré, José
- Page 360 and 361:
340Antonio J. López-Farré, José
- Page 363 and 364:
In: Handbook of Vitamin C Research
- Page 365 and 366:
Vitamin C in the Treatment of Endot
- Page 367 and 368:
Vitamin C in the Treatment of Endot
- Page 369 and 370:
Vitamin C in the Treatment of Endot
- Page 371 and 372:
Vitamin C in the Treatment of Endot
- Page 373 and 374:
Vitamin C in the Treatment of Endot
- Page 375 and 376:
In: Handbook of Vitamin C Research
- Page 377 and 378:
Vitamin C Intake by Japanese Patien
- Page 379 and 380:
Vitamin C Intake by Japanese Patien
- Page 381 and 382:
Vitamin C Intake by Japanese Patien
- Page 383 and 384:
Vitamin C Intake by Japanese Patien
- Page 385 and 386:
Vitamin C Intake by Japanese Patien
- Page 387 and 388:
In: Handbook of Vitamin C Research
- Page 389 and 390:
Reciprocal Effects of Ascorbate on
- Page 391 and 392:
Reciprocal Effects of Ascorbate on
- Page 393 and 394:
Reciprocal Effects of Ascorbate on
- Page 395:
Reciprocal Effects of Ascorbate on
- Page 398 and 399:
378Akihito Ishigamicontrols (12,13)
- Page 400 and 401:
380Akihito IshigamiFigure 3. Vitami
- Page 402 and 403:
382Akihito IshigamiFuture of the SM
- Page 405 and 406:
In: Handbook of Vitamin C Research
- Page 407 and 408:
The Importance of Food Processing o
- Page 409 and 410:
IndexAA , 249abdominal cramps, 243a
- Page 411 and 412:
Index 391atherosclerotic plaque, 23
- Page 413 and 414:
Index 393catalase, 6, 13, 169, 181,
- Page 415 and 416:
Index 395cyclooxygenase-2, 337, 341
- Page 417 and 418:
Index 397endometrium, 286, 294endon
- Page 419 and 420:
Index 399GCS, 18gel, x, 87, 92, 121
- Page 421 and 422:
Index 401hyperglycaemia, 223hypergl
- Page 423 and 424:
Index 403kinetics, 71, 120, 124, 14
- Page 425 and 426:
Index 405metastases, 182metastasis,
- Page 427 and 428:
Index 407nitrate, 14, 88, 179, 234,
- Page 429 and 430:
Index 409phosphodiesterase, 344, 35
- Page 431 and 432:
Index 411recovery, vii, 1, 5, 7, 8,
- Page 433 and 434:
Index 413steady state, 63, 76, 78st
- Page 435:
Index 415UV, 156, 191, 201, 204, 22