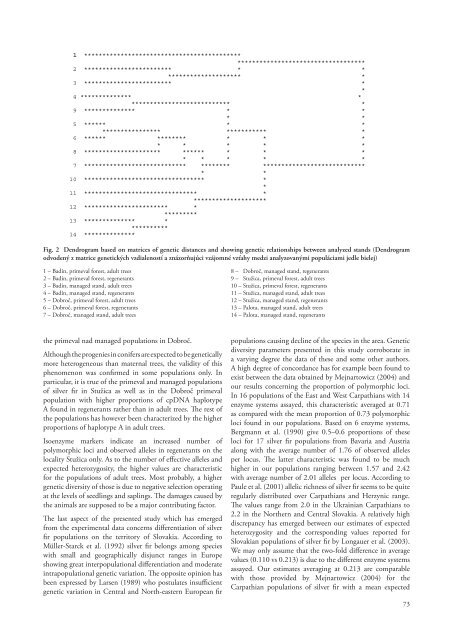
Table 2. Some genetic diversity parameters in three primeval populations of silver fir and in their corresponding managedcounterparts at the levels of adult trees and regenerants (Niektoré ukazovatele genetickej diverzity troch populácií jedle bielejv pralesoch a príslušných obhospodarovaných porastov jedle zistené na úrovni dospelých jedincov a regenerantov)Populations Sample size Polymor. loci N a± st. d. N e± st. d. H e± st. d.Badin – primeval stand, adult trees 68 0.78 1.92 ± 0.61 1.47 ± 0.51 0.250 ± 0.22Badin – primeval stand, regenerants 138 0.78 2.07 ± 0.73 1.39 ± 0.48 0.217 ± 0.20Badin – managed stand, adult trees 60 0.78 1.92 ± 0.61 1.40 ± 0.52 0.216 ± 0.22Badin – managed stand, regenerants 190 0.92 2.42 ± 0.64 1.43 ± 0.53 0.234 ± 0.20Dobroč – primeval stand, adult trees 63 0.57 1.71 ± 0.72 1.35 ± 0.54 0.179 ± 0.22Dobroč – primeval stand, regenerants 78 0.50 1.57 ± 0.64 1.27 ± 0.53 0.135 ± 0.20Dobroč – managed stand, adult trees 80 0.78 2.00 ± 0.67 1.38 ± 0.51 0.208 ± 0.20Dobroč – managed stand, regenerants 126 0.64 1.92 ± 0.82 1.35 ± 0.53 0.188 ± 0.21Stužica – primeval stand, adult trees 81 0.64 1.92 ± 0.82 1.47 ± 0.62 0.227 ± 0.24Stužica – primeval stand, regenerants 115 0.78 2.00 ± 0.67 1.36 ± 0.48 0.194 ± 0.21Stužica – managed stand, adult trees 58 0.71 2.00 ± 0.78 1.40 ± 0.52 0.210 ± 0.22Stužica – managed stand, regenerants 182 0.92 2.28 ± 0.61 1.43 ± 0.53 0.229 ± 0.22Palota – managed stand, adult trees 58 0.64 2.07 ± 0.91 1.55 ± 0.61 0.268 ± 0.24Palota – managed stand, regenerants 176 0.85 2.42 ± 0.75 1.44 ± 0.53 0.238 ± 0.22N a– number of observed alleles, N e– number of effective alleles, H e– expected heterozygositypopulations Badín and Stužica (0.250, 0.227) than in theirmanaged stands (0.216, 0.210). The reverse situation was foundin Dobroč showing higher level of expected heterozygosityin its managed rather than primeval population (0.208 vs0.179 in primeval stand). It is of interest to mention in thisconnection that the highest degree of expected heterozygositywas found in adult trees on the locality Palota (0.268) servingas a managed control to Stužica primeval population. At thelevel of adult trees and regenerated <strong>pro</strong>genies, the formerharbour a higher degree of expected heterozygosity thancorresponding <strong>pro</strong>genies. In particular it is true for all thethree primeval populations analyzed and for the managedpopulations Dobroč and Palota. Managed populations inBadín and Stužica were the only exceptions showing higherdegree of expected heterozygosity in regenerants.Based on analyzed isoenzyme diversity components,a dendrogram was constructed using standard geneticdistances and showing genetic differentiation betweenindividual populations of silver fir (Fig. 2). Their clusteringon the dendrogram reflected both geographic and preservedorigin or managed nature of the populations. The populationsfrom the locality Badín have accordingly been positionedin the upper part of a dendrogram (1–4), while those fromthe locality Stužica in the bottom (10–14). The populationsfrom Dobroč occupy intermediated position (5–8) exhibitinga higher genetic afinity towards populations in Stužica thanto those in Badín. The only exception was in this respect thepopulation of adult trees of Stužica primeval stand (9) whichhas been inserted rather atypically, e. g. between Badín andDobroč populations. It is apparent from a separation on thedendrogram that primeval and managed stands in Badín (1,3) and in Dobroč (5, 7) are <strong>pro</strong>foundly differentiated fromeach other. A common cluster of the managed populations inStužica and those situated 40 km away in Palota may be takenas an evidence supporting their common origin. Contrastingfeature of the illustrated clustering is a short genetic distance72ascertained between adult trees and regenerants in Dobroč (5,6) and extremely large distance between the analogous groupsof trees in Stužica (9, 10) exceeding even the genetic distancesbetween widely separated populations of the species fromdifferent geographic regions of the country. This aspect ofsilver fir genetic differentiation needs to be analyzed further.DISCUSSIONIn Central Europe the silver fir is an integral part of the socalled„Carpatho-Herzynic mixture“ along with Europeanbeach and Norway spruce (Korpeľ, 1995). The „mixture“represents the most <strong>pro</strong>ductive forest ecosystem whichharbours incomparatively greater biodiversity than otherecosystems in the region. According to Paule et al. (2001)the human activity, mainly forest management and industrialpollution, contributed in a variable extent to the narrowingof genetic variability of individual species of the ecosystemaffecting negatively its <strong>pro</strong>ductivity. Only some of the testedmarkers corroborate this conclusion. At the level of cpDNA,it was the hyplotype B which indicates a slight decrease ofits <strong>pro</strong>portion in adult trees of managed populations. Thehaplotype A exhibited an opposite tendency. The same is trueof the isoenzyme polymorphic loci and the observed alleleswhose <strong>pro</strong>portion and number were found to be higher inadult trees of managed rather than in the primeval populations.The most convincing evidence about the negative impactof managing on silver fir populations was <strong>pro</strong>vided by thenumber of effective alleles and expected heterozygosity. As theprincipal components of genetic diversity these parameterswere of higher values in adult trees of the primeval populationsin Badín and Stužica than in their managed counterparts.However, no generalization of the kind can be made asevidenced by the opposite genetic diversity figure observed in
1 ******************************************************************************2 ************************ * ********************* *3 ************************ **4 ************** **************************** *9 ************** * ** *5 ****** * ***************** *********** *6 ****** ******** * * ** * * * *8 ********************* ****** * * ** * * * *7 **************************** ******** ***************************** *10 ********************************* **11 ******************************* *********************12 *********************** **********13 ************** ***********14 **************Fig. 2 Dendrogram based on matrices of genetic distances and showing genetic relationships between analyzed stands (Dendrogramodvodený z matrice genetických vzdialeností a znázorňujúci vzájomné vzťahy medzi analyzovanými populáciami jedle bielej)1 – Badín, primeval forest, adult trees2 – Badín, primeval forest, regenerants3 – Badín, managed stand, adult trees4 – Badín, managed stand, regenerants5 – Dobroč, primeval forest, adult trees6 – Dobroč, primeval forest, regenerants7 – Dobroč, managed stand, adult trees8 – Dobroč, managed stand, regenerants9 – Stužica, primeval forest, adult trees10 – Stužica, primeval forest, regenerants11 – Stužica, managed stand, adult trees12 – Stužica, managed stand, regenerants13 – Palota, managed stand, adult trees14 – Palota, managed stand, regenerantsthe primeval nad managed populations in Dobroč.Although the <strong>pro</strong>genies in conifers are expected to be geneticallymore heterogeneous than maternal trees, the validity of thisphenomenon was confirmed in some populations only. Inparticular, it is true of the primeval and managed populationsof silver fir in Stužica as well as in the Dobroč primevalpopulation with higher <strong>pro</strong>portions of cpDNA haplotypeA found in regenerants rather than in adult trees. The rest ofthe populations has however been characterized by the higher<strong>pro</strong>portions of haplotype A in adult trees.Isoenzyme markers indicate an increased number ofpolymorphic loci and observed alleles in regenerants on thelocality Stužica only. As to the number of effective alleles andexpected heterozygosity, the higher values are characteristicfor the populations of adult trees. Most <strong>pro</strong>bably, a highergenetic diversity of those is due to negative selection operatingat the levels of seedlings and saplings. The damages caused bythe animals are supposed to be a major contributing factor.The last aspect of the presented study which has emergedfrom the experimental data concerns differentiation of silverfir populations on the territory of Slovakia. According toMüller-Starck et al. (1992) silver fir belongs among specieswith small and geographically disjunct ranges in Europeshowing great interpopulational differentiation and moderateintrapopulational genetic variation. The opposite opinion hasbeen expressed by Larsen (1989) who postulates insufficientgenetic variation in Central and North-eastern European firpopulations causing decline of the species in the area. Geneticdiversity parameters presented in this study corroborate ina varying degree the data of these and some other authors.A high degree of concordance has for example been found toexist between the data obtained by Mejnartowicz (2004) andour results concerning the <strong>pro</strong>portion of polymorphic loci.In 16 populations of the East and West Carpathians with 14enzyme systems assayed, this characteristic averaged at 0.71as compared with the mean <strong>pro</strong>portion of 0.73 polymorphicloci found in our populations. Based on 6 enzyme systems,Bergmann et al. (1990) give 0.5–0.6 <strong>pro</strong>portions of theseloci for 17 silver fir populations from Bavaria and Austriaalong with the average number of 1.76 of observed allelesper locus. The latter characteristic was found to be muchhigher in our populations ranging between 1.57 and 2.42with average number of 2.01 alleles per locus. According toPaule et al. (2001) allelic richness of silver fir seems to be quiteregularly distributed over Carpathians and Herzynic range.The values range from 2.0 in the Ukrainian Carpathians to2.2 in the Northern and Central Slovakia. A relatively highdiscrepancy has emerged between our estimates of expectedheterozygosity and the corresponding values reported forSlovakian populations of silver fir by Longauer et al. (2003).We may only assume that the two-fold difference in averagevalues (0.110 vs 0.213) is due to the different enzyme systemsassayed. Our estimates averaging at 0.213 are comparablewith those <strong>pro</strong>vided by Mejnartowicz (2004) for theCarpathian populations of silver fir with a mean expected73
- Page 1 and 2:
A C T AP R U H O N I C I A N A93 20
- Page 3:
OBSAHVliv různých forem železa a
- Page 6 and 7:
(Fe-EDDHA) je z dostupných slouče
- Page 8 and 9:
Tab. 3 Chemické vlastnosti substr
- Page 11 and 12:
Acta Pruhoniciana 93: 11-14, Průho
- Page 13 and 14:
(obr. 3), nebo úzké, protáhlé s
- Page 15 and 16:
Acta Pruhoniciana 93: 15-18, Průho
- Page 17 and 18:
Pro inokulaci byly použity rostlin
- Page 19 and 20:
Acta Pruhoniciana 93: 19-26, Průho
- Page 21 and 22: quite easily. In addition, seedling
- Page 23 and 24: Table 5 Segregation of leaf colour
- Page 25 and 26: Table 7 Percent relative vitality o
- Page 27 and 28: Acta Pruhoniciana 93: 27-30, Průho
- Page 29 and 30: Tab. 1 Rozsah sbírek a možnosti v
- Page 31 and 32: Acta Pruhoniciana 93: 31-35, Průho
- Page 33 and 34: Tab. 2 Vliv koncentrace NAA a cytok
- Page 35: Mitra, G. C. (1989): Biology, conse
- Page 38 and 39: poznatky o našich endemických je
- Page 40 and 41: Sorbus eximia - jeřáb krasovýSor
- Page 42 and 43: lze nalézt habituálně nejlépe v
- Page 44 and 45: 10 Čepel listů okrouhle vejčitá
- Page 46 and 47: Základní taxonomické studium rod
- Page 48 and 49: ný v našich přírodních populac
- Page 50 and 51: ZÁVĚRYPinus neilreichiana je v l
- Page 52 and 53: spěvku je osvětlit ožehavou a v
- Page 54 and 55: cekmenné stromky rozvětvené od z
- Page 56 and 57: začal buk plodit, první silnějš
- Page 59 and 60: Acta Pruhoniciana 93: 59-62, Průho
- Page 61 and 62: Table 1 Characteristics of generati
- Page 63 and 64: Acta Pruhoniciana 93: 63-67, Průho
- Page 65 and 66: nezřídka zaměňovány, přitom n
- Page 67: MSI (2001): Checklist of Magnolia C
- Page 70 and 71: in Eastern Slovakia, on the border
- Page 74 and 75: heterozygosity of 0.269. Also, Konn
- Page 77 and 78: Acta Pruhoniciana 93: 77-82, Průho
- Page 79 and 80: pólové jadrá. Davis (1966) uvád
- Page 81 and 82: Obr. 17 P. spinosa (21. 4. 2009) -
- Page 83 and 84: Acta Pruhoniciana 93: 83-88, Průho
- Page 85 and 86: Na počátku pokusu měly všechny
- Page 87 and 88: Tab. 9 Doronicum orientale ‘Leona
- Page 89 and 90: Acta Pruhoniciana 93: 89-95, Průho
- Page 91 and 92: Popis stanovištěDendrologická za
- Page 93 and 94: Oenothera glazioviana O. erythrosep
- Page 95: preference. Landscape and Urban Pla
- Page 98 and 99: Tab. 1 Původ uloženého osivaDár
- Page 100 and 101: Kapacita regeneračního procesu p
- Page 102 and 103: u nás pěstují, prodávávají ne
- Page 104 and 105: 104Odrůda Šlechtitel Rok Skupina
- Page 106 and 107: 106Odrůda Šlechtitel Rok Skupina
- Page 108 and 109: Výběr odrůd s uvedením cenných
- Page 110 and 111: 110
- Page 112 and 113: Vědecký název Český název Če
- Page 114 and 115: Hottonia palustris L. - žebratka b
- Page 116 and 117: palustris Gaud. na Moravě. Diplomo
- Page 118 and 119: na přírodním stanovišti. Uplatn
- Page 120 and 121: také J. Holzbecher, J. Janouš a V
- Page 122 and 123:
Kultivar Hybridní skupina Původ B
- Page 124 and 125:
Kultivar Hybridní skupina Původ B
- Page 126 and 127:
Kultivar Hybridní skupina Původ B
- Page 128:
Horný, R., Soják, J., Webr, K. M.