Segmentation of Stochastic Images using ... - Jacobs University
Segmentation of Stochastic Images using ... - Jacobs University
Segmentation of Stochastic Images using ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Chapter 5 <strong>Stochastic</strong> <strong>Images</strong><br />
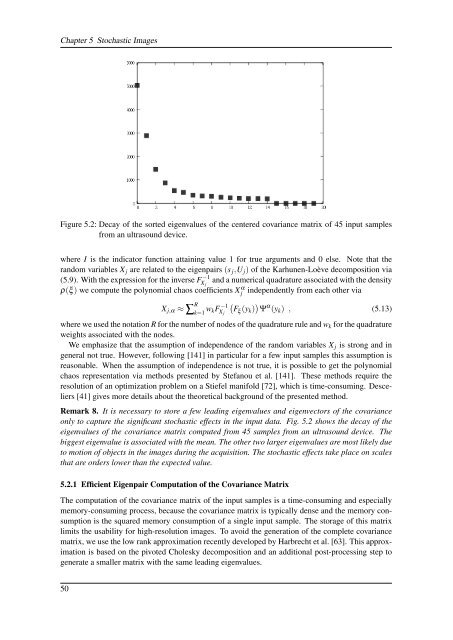
Figure 5.2: Decay <strong>of</strong> the sorted eigenvalues <strong>of</strong> the centered covariance matrix <strong>of</strong> 45 input samples<br />
from an ultrasound device.<br />
where I is the indicator function attaining value 1 for true arguments and 0 else. Note that the<br />
random variables X j are related to the eigenpairs (s j ,U j ) <strong>of</strong> the Karhunen-Loève decomposition via<br />
(5.9). With the expression for the inverse FX −1<br />
j<br />
and a numerical quadrature associated with the density<br />
ρ(ξ ) we compute the polynomial chaos coefficients Xj<br />
α independently from each other via<br />
X j,α ≈ ∑ R k=1 w kF −1<br />
X j<br />
(<br />
Fξ (y k ) ) Ψ α (y k ) , (5.13)<br />
where we used the notation R for the number <strong>of</strong> nodes <strong>of</strong> the quadrature rule and w k for the quadrature<br />
weights associated with the nodes.<br />
We emphasize that the assumption <strong>of</strong> independence <strong>of</strong> the random variables X j is strong and in<br />
general not true. However, following [141] in particular for a few input samples this assumption is<br />
reasonable. When the assumption <strong>of</strong> independence is not true, it is possible to get the polynomial<br />
chaos representation via methods presented by Stefanou et al. [141]. These methods require the<br />
resolution <strong>of</strong> an optimization problem on a Stiefel manifold [72], which is time-consuming. Desceliers<br />
[41] gives more details about the theoretical background <strong>of</strong> the presented method.<br />
Remark 8. It is necessary to store a few leading eigenvalues and eigenvectors <strong>of</strong> the covariance<br />
only to capture the significant stochastic effects in the input data. Fig. 5.2 shows the decay <strong>of</strong> the<br />
eigenvalues <strong>of</strong> the covariance matrix computed from 45 samples from an ultrasound device. The<br />
biggest eigenvalue is associated with the mean. The other two larger eigenvalues are most likely due<br />
to motion <strong>of</strong> objects in the images during the acquisition. The stochastic effects take place on scales<br />
that are orders lower than the expected value.<br />
5.2.1 Efficient Eigenpair Computation <strong>of</strong> the Covariance Matrix<br />
The computation <strong>of</strong> the covariance matrix <strong>of</strong> the input samples is a time-consuming and especially<br />
memory-consuming process, because the covariance matrix is typically dense and the memory consumption<br />
is the squared memory consumption <strong>of</strong> a single input sample. The storage <strong>of</strong> this matrix<br />
limits the usability for high-resolution images. To avoid the generation <strong>of</strong> the complete covariance<br />
matrix, we use the low rank approximation recently developed by Harbrecht et al. [63]. This approximation<br />
is based on the pivoted Cholesky decomposition and an additional post-processing step to<br />
generate a smaller matrix with the same leading eigenvalues.<br />
50