Segmentation of Stochastic Images using ... - Jacobs University
Segmentation of Stochastic Images using ... - Jacobs University
Segmentation of Stochastic Images using ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
5.2 Generation <strong>of</strong> <strong>Stochastic</strong> <strong>Images</strong> from Samples<br />
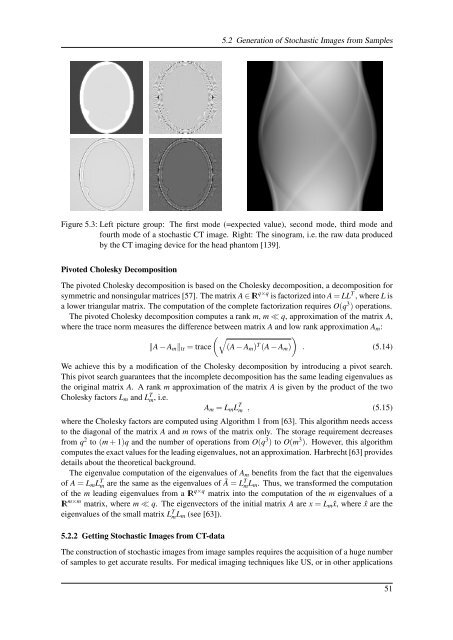
Figure 5.3: Left picture group: The first mode (=expected value), second mode, third mode and<br />
fourth mode <strong>of</strong> a stochastic CT image. Right: The sinogram, i.e. the raw data produced<br />
by the CT imaging device for the head phantom [139].<br />
Pivoted Cholesky Decomposition<br />
The pivoted Cholesky decomposition is based on the Cholesky decomposition, a decomposition for<br />
symmetric and nonsingular matrices [57]. The matrix A ∈ IR q×q is factorized into A = LL T , where L is<br />
a lower triangular matrix. The computation <strong>of</strong> the complete factorization requires O(q 3 ) operations.<br />
The pivoted Cholesky decomposition computes a rank m, m ≪ q, approximation <strong>of</strong> the matrix A,<br />
where the trace norm measures the difference between matrix A and low rank approximation A m :<br />
(√<br />
)<br />
‖A − A m ‖ tr = trace (A − A m ) T (A − A m ) . (5.14)<br />
We achieve this by a modification <strong>of</strong> the Cholesky decomposition by introducing a pivot search.<br />
This pivot search guarantees that the incomplete decomposition has the same leading eigenvalues as<br />
the original matrix A. A rank m approximation <strong>of</strong> the matrix A is given by the product <strong>of</strong> the two<br />
Cholesky factors L m and L T m, i.e.<br />
A m = L m L T m , (5.15)<br />
where the Cholesky factors are computed <strong>using</strong> Algorithm 1 from [63]. This algorithm needs access<br />
to the diagonal <strong>of</strong> the matrix A and m rows <strong>of</strong> the matrix only. The storage requirement decreases<br />
from q 2 to (m + 1)q and the number <strong>of</strong> operations from O(q 3 ) to O(m 3 ). However, this algorithm<br />
computes the exact values for the leading eigenvalues, not an approximation. Harbrecht [63] provides<br />
details about the theoretical background.<br />
The eigenvalue computation <strong>of</strong> the eigenvalues <strong>of</strong> A m benefits from the fact that the eigenvalues<br />
<strong>of</strong> A = L m L T m are the same as the eigenvalues <strong>of</strong> Ã = L T mL m . Thus, we transformed the computation<br />
<strong>of</strong> the m leading eigenvalues from a IR q×q matrix into the computation <strong>of</strong> the m eigenvalues <strong>of</strong> a<br />
IR m×m matrix, where m ≪ q. The eigenvectors <strong>of</strong> the initial matrix A are x = L m ˆx, where ˆx are the<br />
eigenvalues <strong>of</strong> the small matrix L T mL m (see [63]).<br />
5.2.2 Getting <strong>Stochastic</strong> <strong>Images</strong> from CT-data<br />
The construction <strong>of</strong> stochastic images from image samples requires the acquisition <strong>of</strong> a huge number<br />
<strong>of</strong> samples to get accurate results. For medical imaging techniques like US, or in other applications<br />
51