You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
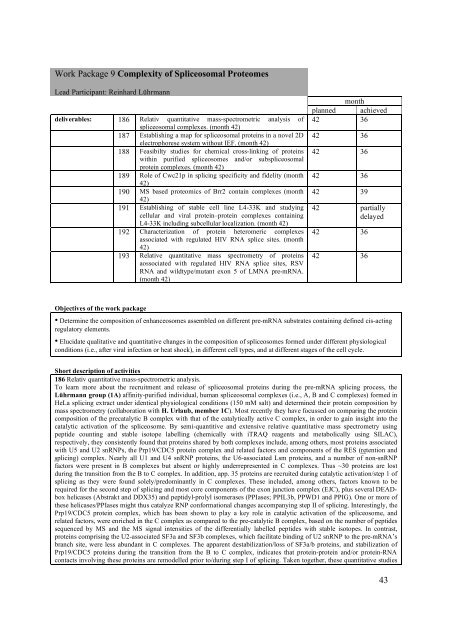
Work Package 9 Complexity of Spliceosomal ProteomesLead Participant: Reinhard Lührmanndeliverables: 186 Relativ quantitative mass-spectrometric analysis ofspliceosomal complexes. (month 42)187 Establishing a map for spliceosomal proteins in a novel 2Delectrophorese system without IEF. (month 42)188 Feasibilty studies for chemical cross-linking of proteinswithin purified spliceosomes and/or subspliceosomalprotein complexes. (month 42)189 Role of Cwc21p in splicing specificity and fidelity (month42)190 MS based proteomics of Brr2 contain complexes (month42)191 Establishing of stable cell line L4-33K and studyingcellular and viral protein–protein complexes containingL4-33K including subcellular localization. (month 42)192 Characterization of protein heteromeric complexesassociated with regulated HIV RNA splice sites. (month42)193 Relative quantitative mass spectrometry of proteinsaossociated with regulated HIV RNA splice sites, RSVRNA and wildtype/mutant exon 5 of LMNA pre-mRNA.(month 42)monthplanned achieved42 3642 3642 3642 3642 3942 partiallydelayed42 3642 36Objectives of the work package• Determine the composition of enhanceosomes assembled on different pre-mRNA substrates containing defined cis-actingregulatory elements.• Elucidate qualitative and quantitative changes in the composition of spliceosomes formed under different physiologicalconditions (i.e., after viral infection or heat shock), in different cell types, and at different stages of the cell cycle.Short description of activities186 Relativ quantitative mass-spectrometric analysis.To learn more about the recruitment and release of spliceosomal proteins during the pre-mRNA splicing process, theLührmann group (1A) affinity-purified individual, human spliceosomal complexes (i.e., A, B and C complexes) formed inHeLa splicing extract under identical physiological conditions (150 mM salt) and determined their protein composition bymass spectrometry (collaboration with H. Urlaub, member 1C). Most recently they have focussed on comparing the proteincomposition of the precatalytic B complex with that of the catalytically active C complex, in order to gain insight into thecatalytic activation of the spliceosome. By semi-quantitive and extensive relative quantitative mass spectrometry usingpeptide counting and stable isotope labelling (chemically with iTRAQ reagents and metabolically using SILAC),respectively, they consistently found that proteins shared by both complexes include, among others, most proteins associatedwith U5 and U2 snRNPs, the Prp19/CDC5 protein complex and related factors and components of the RES (retention andsplicing) complex. Nearly all U1 and U4 snRNP proteins, the U6-associated Lsm proteins, and a number of non-snRNPfactors were present in B complexes but absent or highly underrepresented in C complexes. Thus ~30 proteins are lostduring the transition from the B to C complex. In addition, app. 35 proteins are recruited during catalytic activation/step 1 ofsplicing as they were found solely/predominantly in C complexes. These included, among others, factors known to berequired for the second step of splicing and most core components of the exon junction complex (EJC), plus several DEADboxhelicases (Abstrakt and DDX35) and peptidyl-prolyl isomerases (PPIases; PPIL3b, PPWD1 and PPIG). One or more ofthese helicases/PPIases might thus catalyze RNP conformational changes accompanying step II of splicing. Interestingly, thePrp19/CDC5 protein complex, which has been shown to play a key role in catalytic activation of the spliceosome, andrelated factors, were enriched in the C complex as compared to the pre-catalytic B complex, based on the number of peptidessequenced by MS and the MS signal intensities of the differentially labelled peptides with stable isotopes. In contrast,proteins comprising the U2-associated SF3a and SF3b complexes, which facilitate binding of U2 snRNP to the pre-mRNA’sbranch site, were less abundant in C complexes. The apparent destabilization/loss of SF3a/b proteins, and stabilization ofPrp19/CDC5 proteins during the transition from the B to C complex, indicates that protein-protein and/or protein-RNAcontacts involving these proteins are remodelled prior to/during step I of splicing. Taken together, these quantitative studies43