Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
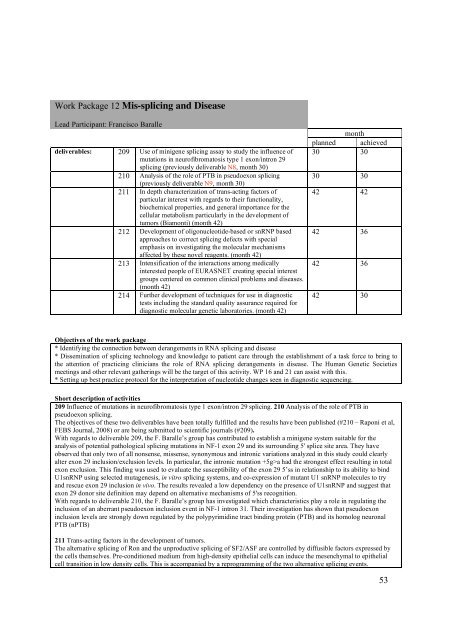
Work Package 12 Mis-splicing and DiseaseLead Participant: Francisco Baralledeliverables: 209 Use of minigene splicing assay to study the influence ofmutations in neurofibromatosis type 1 exon/intron 29splicing (previously deliverable N8, month 30)210 Analysis of the role of PTB in pseudoexon splicing(previously deliverable N9, month 30)211 In depth characterization of trans-acting factors ofparticular interest with regards to their functionality,biochemical properties, and general importance for thecellular metabolism particularly in the development oftumors (Biamonti) (month 42)212 Development of oligonucleotide-based or snRNP basedapproaches to correct splicing defects with specialemphasis on investigating the molecular mechanismsaffected by these novel reagents. (month 42)213 Intensification of the interactions among medicallyinterested people of <strong>EURASNET</strong> creating special interestgroups centered on common clinical problems and diseases.(month 42)214 Further development of techniques for use in diagnostictests including the standard quality assurance required fordiagnostic molecular genetic laboratories. (month 42)monthplanned achieved30 3030 3042 4242 3642 3642 30Objectives of the work package* Identifying the connection between derangements in RNA splicing and disease* Dissemination of splicing technology and knowledge to patient care through the establishment of a task force to bring tothe attention of practicing clinicians the role of RNA splicing derangements in disease. The Human Genetic Societiesmeetings and other relevant gatherings will be the target of this activity. WP 16 and 21 can assist with this.* Setting up best practice protocol for the interpretation of nucleotide changes seen in diagnostic sequencing.Short description of activities209 Influence of mutations in neurofibromatosis type 1 exon/intron 29 splicing. 210 Analysis of the role of PTB inpseudoexon splicing.The objectives of these two deliverables have been totally fulfilled and the results have been published (#210 – Raponi et al,FEBS Journal, <strong>2008</strong>) or are being submitted to scientific journals (#209).With regards to deliverable 209, the F. Baralle’s group has contributed to establish a minigene system suitable for theanalysis of potential pathological splicing mutations in NF-1 exon 29 and its surrounding 5' splice site area. They haveobserved that only two of all nonsense, missense, synonymous and intronic variations analyzed in this study could clearlyalter exon 29 inclusion/exclusion levels. In particular, the intronic mutation +5g>a had the strongest effect resulting in totalexon exclusion. This finding was used to evaluate the susceptibility of the exon 29 5’ss in relationship to its ability to bindU1snRNP using selected mutagenesis, in vitro splicing systems, and co-expression of mutant U1 snRNP molecules to tryand rescue exon 29 inclusion in vivo. The results revealed a low dependency on the presence of U1snRNP and suggest thatexon 29 donor site definition may depend on alternative mechanisms of 5'ss recognition.With regards to deliverable 210, the F. Baralle’s group has investigated which characteristics play a role in regulating theinclusion of an aberrant pseudoexon inclusion event in NF-1 intron 31. Their investigation has shown that pseudoexoninclusion levels are strongly down regulated by the polypyrimidine tract binding protein (PTB) and its homolog neuronalPTB (nPTB)211 Trans-acting factors in the development of tumors.The alternative splicing of Ron and the unproductive splicing of SF2/ASF are controlled by diffusible factors expressed bythe cells themselves. Pre-conditioned medium from high-density epithelial cells can induce the mesenchymal to epithelialcell transition in low density cells. This is accompanied by a reprogramming of the two alternative splicing events.53