Sequencher 4.8 User Manual--PC - Bioinformatics and Biological ...
Sequencher 4.8 User Manual--PC - Bioinformatics and Biological ...
Sequencher 4.8 User Manual--PC - Bioinformatics and Biological ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
When you click the Show Chromatograms button in a region without trace data<br />
you will see a “No Chromatogram Data” message in the Contig Chromatogram<br />
window. When you move the cursor back into a region where there are fragments with<br />
chromatogram data, the traces will reappear.<br />
Underst<strong>and</strong>ing the Trace Display<br />
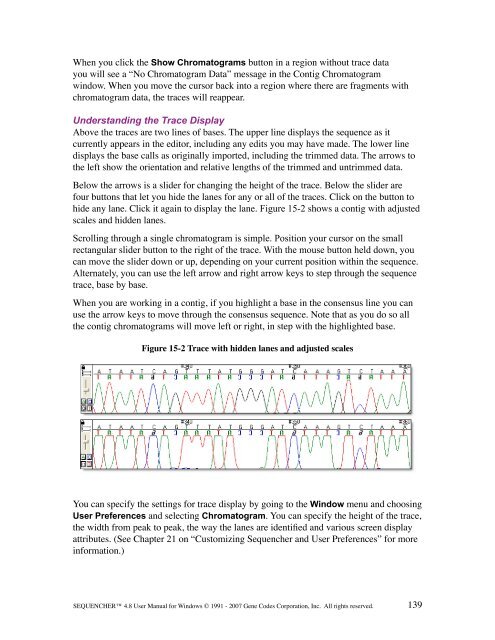
Above the traces are two lines of bases. The upper line displays the sequence as it<br />
currently appears in the editor, including any edits you may have made. The lower line<br />
displays the base calls as originally imported, including the trimmed data. The arrows to<br />
the left show the orientation <strong>and</strong> relative lengths of the trimmed <strong>and</strong> untrimmed data.<br />
Below the arrows is a slider for changing the height of the trace. Below the slider are<br />
four buttons that let you hide the lanes for any or all of the traces. Click on the button to<br />
hide any lane. Click it again to display the lane. Figure 15-2 shows a contig with adjusted<br />
scales <strong>and</strong> hidden lanes.<br />
Scrolling through a single chromatogram is simple. Position your cursor on the small<br />
rectangular slider button to the right of the trace. With the mouse button held down, you<br />
can move the slider down or up, depending on your current position within the sequence.<br />
Alternately, you can use the left arrow <strong>and</strong> right arrow keys to step through the sequence<br />
trace, base by base.<br />
When you are working in a contig, if you highlight a base in the consensus line you can<br />
use the arrow keys to move through the consensus sequence. Note that as you do so all<br />
the contig chromatograms will move left or right, in step with the highlighted base.<br />
Figure 15-2 Trace with hidden lanes <strong>and</strong> adjusted scales<br />
You can specify the settings for trace display by going to the Window menu <strong>and</strong> choosing<br />
<strong>User</strong> Preferences <strong>and</strong> selecting Chromatogram. You can specify the height of the trace,<br />
the width from peak to peak, the way the lanes are identified <strong>and</strong> various screen display<br />
attributes. (See Chapter 21 on “Customizing <strong>Sequencher</strong> <strong>and</strong> <strong>User</strong> Preferences” for more<br />
information.)<br />
SEQUENCHER <strong>4.8</strong> <strong>User</strong> <strong>Manual</strong> for Windows © 1991 - 2007 Gene Codes Corporation, Inc. All rights reserved.<br />
139