Sequencher 4.8 User Manual--PC - Bioinformatics and Biological ...
Sequencher 4.8 User Manual--PC - Bioinformatics and Biological ...
Sequencher 4.8 User Manual--PC - Bioinformatics and Biological ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Interactive assembly<br />
The Assemble Interactively algorithm can be used whenever you want complete<br />
control over the assembly of a batch of sequences. The Assemble Interactively<br />
window provides you with detailed information regarding overlap, mismatch <strong>and</strong><br />
gapping for any given sequence <strong>and</strong> a set of c<strong>and</strong>idate sequences. The c<strong>and</strong>idate<br />
assemblies are driven by your user-defined assembly parameter settings.<br />
Performing an interactive assembly<br />
Bring the Project Window to the front <strong>and</strong> select the items you want to assemble.<br />
Click on the Assemble Interactively button at the top of the Project Window or go to<br />
the Contig menu <strong>and</strong> choose Assemble Contigs <strong>and</strong> then Interactively… from the<br />
submenu.<br />
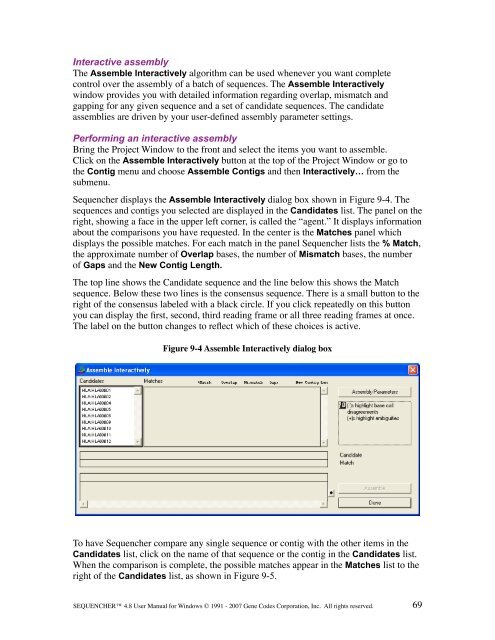
<strong>Sequencher</strong> displays the Assemble Interactively dialog box shown in Figure 9-4. The<br />
sequences <strong>and</strong> contigs you selected are displayed in the C<strong>and</strong>idates list. The panel on the<br />
right, showing a face in the upper left corner, is called the “agent.” It displays information<br />
about the comparisons you have requested. In the center is the Matches panel which<br />
displays the possible matches. For each match in the panel <strong>Sequencher</strong> lists the % Match,<br />
the approximate number of Overlap bases, the number of Mismatch bases, the number<br />
of Gaps <strong>and</strong> the New Contig Length.<br />
The top line shows the C<strong>and</strong>idate sequence <strong>and</strong> the line below this shows the Match<br />
sequence. Below these two lines is the consensus sequence. There is a small button to the<br />
right of the consensus labeled with a black circle. If you click repeatedly on this button<br />
you can display the first, second, third reading frame or all three reading frames at once.<br />
The label on the button changes to reflect which of these choices is active.<br />
Figure 9-4 Assemble Interactively dialog box<br />
To have <strong>Sequencher</strong> compare any single sequence or contig with the other items in the<br />
C<strong>and</strong>idates list, click on the name of that sequence or the contig in the C<strong>and</strong>idates list.<br />
When the comparison is complete, the possible matches appear in the Matches list to the<br />
right of the C<strong>and</strong>idates list, as shown in Figure 9-5.<br />
SEQUENCHER <strong>4.8</strong> <strong>User</strong> <strong>Manual</strong> for Windows © 1991 - 2007 Gene Codes Corporation, Inc. All rights reserved.<br />
69