- Page 2 and 3: THIS IS AN AGREEMENT BETWEEN YOU, T
- Page 4 and 5: © 1991 - 2007 Gene Codes Corporati
- Page 6 and 7: Templates .........................
- Page 8 and 9: Set Circular Genome Size ..........
- Page 10 and 11: The Bases View ....................
- Page 12 and 13: Protein translations for the Consen
- Page 14 and 15: Setting header, footer and margin o
- Page 16 and 17: Page Intentionally Left Blank xv
- Page 18 and 19: We welcome your comments or support
- Page 20 and 21: Navigating Sequencher Help The Sequ
- Page 22 and 23: Contig menu The commands in the Con
- Page 24 and 25: Page Intentionally Left Blank SEQUE
- Page 26 and 27: Figure 3-1 An empty Sequencher Proj
- Page 28 and 29: Figure 3-4 Project Window with Larg
- Page 30 and 31: value in the Confidence pane of the
- Page 32 and 33: Editing the information for a seque
- Page 34 and 35: Page Intentionally Left Blank SEQUE
- Page 36 and 37: How to import using menu commands I
- Page 38 and 39: Figure 4-2 Browse for Folder dialog
- Page 40 and 41: Importing Confidence Scores Many ba
- Page 42 and 43: If you have finished working with S
- Page 44 and 45: Selecting sequences Selecting seque
- Page 46 and 47: Renaming a sequence or contig Renam
- Page 48 and 49: Closing the Project Window When you
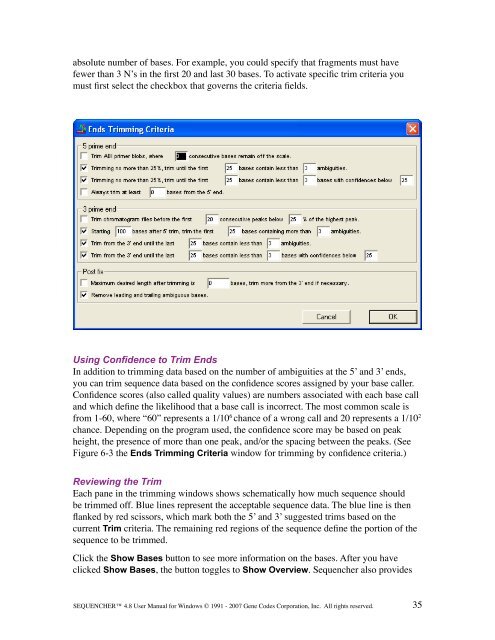
- Page 52 and 53: additional control within the Trim
- Page 54 and 55: Figure 6-7 The polylinker window No
- Page 56 and 57: Setting Vector Trimming Criteria Se
- Page 58 and 59: Page Intentionally Left Blank SEQUE
- Page 60 and 61: Basic Reference Sequence properties
- Page 62 and 63: When the To Reference by Name comma
- Page 64 and 65: Page Intentionally Left Blank SEQUE
- Page 66 and 67: A locked editor is read-only. You c
- Page 68 and 69: If your sequence was called “MyPU
- Page 70 and 71: Viewing summary information in the
- Page 72 and 73: Figure 8-7 ORF selected by Next Met
- Page 74 and 75: If you want to assign a personal an
- Page 76 and 77: Voice verification Sequencher provi
- Page 78 and 79: Page Intentionally Left Blank SEQUE
- Page 80 and 81: Chapter 9 “Sequence Assembly” f
- Page 82 and 83: Figure 9-2 The Clean Data algorithm
- Page 84 and 85: Performing the assembly Automatic a
- Page 86 and 87: Figure 9-5 Interactive assembly sho
- Page 88 and 89: The new contig will appear in the P
- Page 90 and 91: When you use a Name Delimiter, it w
- Page 92 and 93: If the Expression is a delimiter bo
- Page 94 and 95: Figure 10-6 The Project Window with
- Page 96 and 97: Page Intentionally Left Blank SEQUE
- Page 98 and 99: it is in the reverse orientation. T
- Page 100 and 101:
Figure 11-2 Contig overview Fifure
- Page 102 and 103:
Base Numbers At Transitions Clickin
- Page 104 and 105:
Label Name Function E F G H Sequenc
- Page 106 and 107:
Consensus to Forensic Standards In
- Page 108 and 109:
consensus is from a base with a low
- Page 110 and 111:
move the gaps to the 5’ end, sele
- Page 112 and 113:
Figure 12-4 Fill Void dialog box De
- Page 114 and 115:
First select the base or range of b
- Page 116 and 117:
Page Intentionally Left Blank SEQUE
- Page 118 and 119:
The Translated Variance Table If yo
- Page 120 and 121:
Structure of the Translated Varianc
- Page 122 and 123:
(see Figure 13-5). It is possible f
- Page 124 and 125:
Figure 13-7 Translated Variance Tab
- Page 126 and 127:
If you want to restrict the Compari
- Page 128 and 129:
Figure 13-13 Creating a Joined feat
- Page 130 and 131:
In either case, the method you choo
- Page 132 and 133:
You can edit your data directly in
- Page 134 and 135:
Figure 13-17 The Translated Varianc
- Page 136 and 137:
Figure 13-19 Extract from Variance
- Page 138 and 139:
The Population Table lists the vari
- Page 140 and 141:
If you want to remove sample sequen
- Page 142 and 143:
Select Individual Variance Reports
- Page 144 and 145:
If you prefer to copy your Translat
- Page 146 and 147:
You can copy your Translated Varian
- Page 148 and 149:
Summary report display options Sequ
- Page 150 and 151:
Enabling the protein display To ena
- Page 152 and 153:
Page Intentionally Left Blank SEQUE
- Page 154 and 155:
Note: Remember that even if a seque
- Page 156 and 157:
If you select the Contig Chromatogr
- Page 158 and 159:
Select command Table 15-1 Summary o
- Page 160 and 161:
Note: The Extend Selection command
- Page 162 and 163:
Figure 16-2 Restriction map options
- Page 164 and 165:
example, if you were displaying onl
- Page 166 and 167:
Figure 16-8 Checkmarked enzyme list
- Page 168 and 169:
display this view by going to the V
- Page 170 and 171:
The default setting shows exact mat
- Page 172 and 173:
Creating and editing features To cr
- Page 174 and 175:
Quick feature creation To create a
- Page 176 and 177:
Page Intentionally Left Blank SEQUE
- Page 178 and 179:
Figure 18-1 A dimmed icon Finding t
- Page 180 and 181:
Note: If you pause between characte
- Page 182 and 183:
a dialog box (Figure 18-6). This le
- Page 184 and 185:
Page Intentionally Left Blank SEQUE
- Page 186 and 187:
Figure 19-1 Export Options dialog b
- Page 188 and 189:
You should remember to provide a re
- Page 190 and 191:
Figure 19-3 Copy As Protein Transla
- Page 192 and 193:
[Project Name] and [User Name]. You
- Page 194 and 195:
Page Intentionally Left Blank SEQUE
- Page 196 and 197:
Colors as Backgrounds Sometimes, di
- Page 198 and 199:
Choosing a genetic code You will fi
- Page 200 and 201:
time you select the Review button f
- Page 202 and 203:
Label & Name The Label & Name prefe
- Page 204 and 205:
Click the Use Blank Project radio b
- Page 206 and 207:
Use the Columns For Multiple Traces
- Page 208 and 209:
If you are making changes to the wa
- Page 210 and 211:
Variance Table The Variance Table p
- Page 212 and 213:
Page Intentionally Left Blank SEQUE
- Page 214 and 215:
Figure 22-1 The Validate mtDNA Prof
- Page 216 and 217:
Figure 22-3 The Circular Genome Siz
- Page 218 and 219:
Page Intentionally Left Blank SEQUE
- Page 220 and 221:
Menu/Window Command Windows Select
- Page 222 and 223:
You may require a more complex deli
- Page 224 and 225:
Page Intentionally Left Blank SEQUE
- Page 226 and 227:
Code Base(s) Meaning 5 A or Another
- Page 228 and 229:
Feature Display Color Style Second
- Page 230 and 231:
Page Intentionally Left Blank SEQUE
- Page 232 and 233:
eferences numbers, sequences, or us
- Page 234 and 235:
Qualifier type Description Citation
- Page 236 and 237:
Feature Key Default Feature Qualifi
- Page 238 and 239:
Page Intentionally Left Blank SEQUE
- Page 240 and 241:
Name CursorProject Window Dialog bo