Diseases and Management of Crops under Protected Cultivation
Diseases and Management of Crops under Protected Cultivation
Diseases and Management of Crops under Protected Cultivation
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
(<strong>Diseases</strong> <strong>and</strong> <strong>Management</strong> <strong>of</strong> <strong>Crops</strong> <strong>under</strong> <strong>Protected</strong> <strong>Cultivation</strong>)<br />
dependent on the performance on the PCR assay itself, but also on the efficiency <strong>of</strong> the procedure<br />
employed to extract the nucleic acids from the plant material. To check for substances that may<br />
interfere with the amplification process, internal controls can be designed for each pair <strong>of</strong> primers,<br />
or real-time PCR can be employed. PCR efficiency is controlled by many parameters, such as<br />
polymerase type, buffer composition <strong>and</strong> stability, purity <strong>and</strong> concentration <strong>of</strong> dNTPs, cycling<br />
parameters as well as the characteristics <strong>of</strong> the starting template. Several expensive commercial<br />
integrated systems allow for the automated extraction <strong>and</strong> analysis <strong>of</strong> nucleic acids from<br />
microorganisms, but they are not efficient with all types <strong>of</strong> plant material <strong>and</strong> need to be evaluated<br />
before they can be adopted for routine detection. The specific primers have been designed based<br />
on either the amplification <strong>of</strong> specific genes from the chromosome or plasmids ( Table 3) or on<br />
different approaches such as sequences selected from RAPD differential b<strong>and</strong>s obtained by<br />
subtractive hybridization. Some specific primers have been developed for detection <strong>of</strong> R.<br />
solanacearum from tuber, seeds <strong>and</strong> planting material, soil as well as irrigation is given in Table 2<br />
& 3.<br />
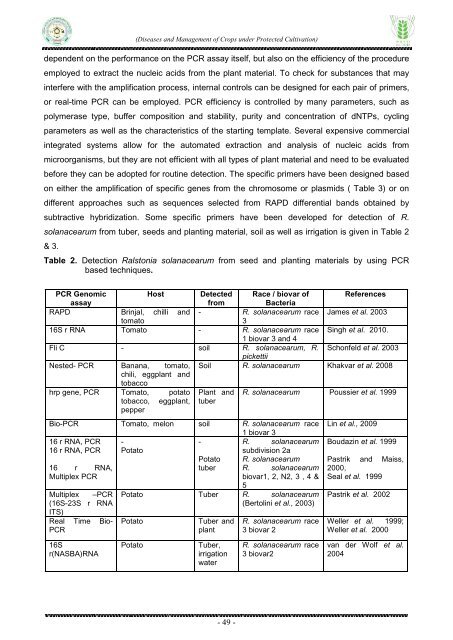
Table 2. Detection Ralstonia solanacearum from seed <strong>and</strong> planting materials by using PCR<br />
based techniques.<br />
PCR Genomic<br />
Host Detected Race / biovar <strong>of</strong> References<br />
assay<br />
from<br />
Bacteria<br />
RAPD Brinjal, chilli <strong>and</strong> - R. solanacearum race James et al. 2003<br />
tomato<br />
3<br />
16S r RNA Tomato - R. solanacearum race Singh et al. 2010.<br />
1 biovar 3 <strong>and</strong> 4<br />
Fli C - soil R. solanacearum, R. Schonfeld et al. 2003<br />
pickettii<br />
Nested- PCR Banana, tomato, Soil R. solanacearum Khakvar et al. 2008<br />
chili, eggplant <strong>and</strong><br />
tobacco<br />
hrp gene, PCR Tomato, potato Plant <strong>and</strong> R. solanacearum Poussier et al. 1999<br />
tobacco, eggplant, tuber<br />
pepper<br />
Bio-PCR Tomato, melon soil R. solanacearum race<br />
1 biovar 3<br />
16 r RNA, PCR -<br />
-<br />
16 r RNA, PCR Potato<br />
16 r RNA,<br />
Multiplex PCR<br />
Multiplex –PCR<br />
(16S-23S r RNA<br />
ITS)<br />
Real Time Bio-<br />
PCR<br />
Potato<br />
tuber<br />
R. solanacearum<br />
subdivision 2a<br />
R. solanacearum<br />
R. solanacearum<br />
biovar1, 2, N2, 3 , 4 &<br />
5<br />
Potato Tuber R. solanacearum<br />
(Bertolini et al., 2003)<br />
Potato<br />
Tuber <strong>and</strong><br />
plant<br />
R. solanacearum race<br />
3 biovar 2<br />
Lin et al., 2009<br />
Boudazin et al. 1999<br />
Pastrik <strong>and</strong> Maiss,<br />
2000,<br />
Seal et al. 1999<br />
Pastrik et al. 2002<br />
Weller et al. 1999;<br />
Weller et al. 2000<br />
16S<br />
r(NASBA)RNA<br />
Potato<br />
Tuber,<br />
irrigation<br />
water<br />
R. solanacearum race<br />
3 biovar2<br />
van der Wolf et al.<br />
2004<br />
- 49 -