Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
Colletotrichum: complex species or species ... - CBS - KNAW
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
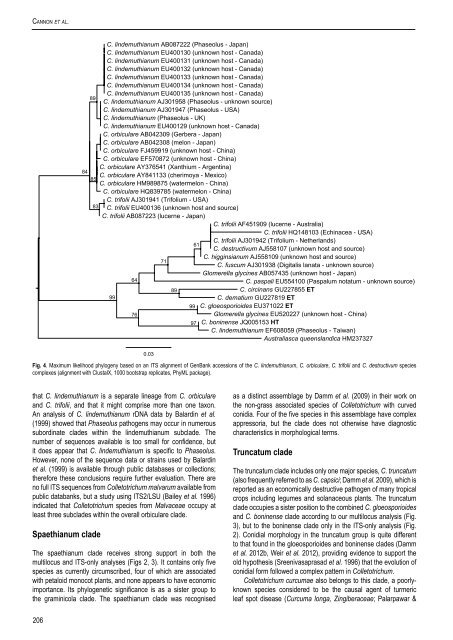
Cannon et al.C. lindemuthianum AB087222 (Phaseolus - Japan)C. lindemuthianum EU400130 (unknown host - Canada)C. lindemuthianum EU400131 (unknown host - Canada)C. lindemuthianum EU400132 (unknown host - Canada)C. lindemuthianum EU400133 (unknown host - Canada)C. lindemuthianum EU400134 (unknown host - Canada)C. lindemuthianum EU400135 (unknown host - Canada)89C. lindemuthianum AJ301958 (Phaseolus - unknown source)C. lindemuthianum AJ301947 (Phaseolus - USA)C. lindemuthianum (Phaseolus - UK)C. lindemuthianum EU400129 (unknown host - Canada)C. <strong>or</strong>biculare AB042309 (Gerbera - Japan)C. <strong>or</strong>biculare AB042308 (melon - Japan)C. <strong>or</strong>biculare FJ459919 (unknown host - China)C. <strong>or</strong>biculare EF570872 (unknown host - China)C. <strong>or</strong>biculare AY376541 (Xanthium - Argentina)84C. <strong>or</strong>biculare AY841133 (cherimoya - Mexico)85C. <strong>or</strong>biculare HM989875 (watermelon - China)C. <strong>or</strong>biculare HQ839785 (watermelon - China)C. trifolii AJ301941 (Trifolium - USA)83 C. trifolii EU400136 (unknown host and source)C. trifolii AB087223 (lucerne - Japan)C. trifolii AF451909 (lucerne - Australia)C. trifolii HQ148103 (Echinacea - USA)C. trifolii AJ301942 (Trifolium - Netherlands)61C. destructivum AJ558107 (unknown host and source)C. higginsianum AJ558109 (unknown host and source)71C. fuscum AJ301938 (Digitalis lanata - unknown source)Glomerella glycines AB057435 (unknown host - Japan)64C. paspali EU554100 (Paspalum notatum - unknown source)89C. circinans GU227855 ET99C. dematium GU227819 ET99 C. gloeosp<strong>or</strong>ioides EU371022 ET76Glomerella glycines EU520227 (unknown host - China)97 C. boninense JQ005153 HTC. lindemuthianum EF608059 (Phaseolus - Taiwan)Australiasca queenslandica HM2373270.03Fig. 4. Maximum likelihood phylogeny based on an ITS alignment of GenBank accessions of the C. lindemuthianum, C. <strong>or</strong>biculare, C. trifolii and C. destructivum <strong>species</strong><strong>complex</strong>es (alignment with ClustalX, 1000 bootstrap replicates, PhyML package).that C. lindemuthianum is a separate lineage from C. <strong>or</strong>biculareand C. trifolii, and that it might comprise m<strong>or</strong>e than one taxon.An analysis of C. lindemuthianum rDNA data by Balardin et al.(1999) showed that Phaseolus pathogens may occur in numeroussub<strong>or</strong>dinate clades within the lindemuthianum subclade. Thenumber of sequences available is too small f<strong>or</strong> confidence, butit does appear that C. lindemuthianum is specific to Phaseolus.However, none of the sequence data <strong>or</strong> strains used by Balardinet al. (1999) is available through public databases <strong>or</strong> collections;theref<strong>or</strong>e these conclusions require further evaluation. There areno full ITS sequences from <strong>Colletotrichum</strong> malvarum available frompublic databanks, but a study using ITS2/LSU (Bailey et al. 1996)indicated that <strong>Colletotrichum</strong> <strong>species</strong> from Malvaceae occupy atleast three subclades within the overall <strong>or</strong>biculare clade.Spaethianum cladeThe spaethianum clade receives strong supp<strong>or</strong>t in both themultilocus and ITS-only analyses (Figs 2, 3). It contains only five<strong>species</strong> as currently circumscribed, four of which are associatedwith petaloid monocot plants, and none appears to have economicimp<strong>or</strong>tance. Its phylogenetic significance is as a sister group tothe graminicola clade. The spaethianum clade was recognisedas a distinct assemblage by Damm et al. (2009) in their w<strong>or</strong>k onthe non-grass associated <strong>species</strong> of <strong>Colletotrichum</strong> with curvedconidia. Four of the five <strong>species</strong> in this assemblage have <strong>complex</strong>appress<strong>or</strong>ia, but the clade does not otherwise have diagnosticcharacteristics in m<strong>or</strong>phological terms.Truncatum cladeThe truncatum clade includes only one maj<strong>or</strong> <strong>species</strong>, C. truncatum(also frequently referred to as C. capsici; Damm et al. 2009), which isrep<strong>or</strong>ted as an economically destructive pathogen of many tropicalcrops including legumes and solanaceous plants. The truncatumclade occupies a sister position to the combined C. gloeosp<strong>or</strong>ioidesand C. boninense clade acc<strong>or</strong>ding to our multilocus analysis (Fig.3), but to the boninense clade only in the ITS-only analysis (Fig.2). Conidial m<strong>or</strong>phology in the truncatum group is quite differentto that found in the gloeosp<strong>or</strong>ioides and boninense clades (Dammet al. 2012b, Weir et al. 2012), providing evidence to supp<strong>or</strong>t theold hypothesis (Sreenivasaprasad et al. 1996) that the evolution ofconidial f<strong>or</strong>m followed a <strong>complex</strong> pattern in <strong>Colletotrichum</strong>.<strong>Colletotrichum</strong> curcumae also belongs to this clade, a po<strong>or</strong>lyknown<strong>species</strong> considered to be the causal agent of turmericleaf spot disease (Curcuma longa, Zingiberaceae; Palarpawar &206