Windows QTL Cartographer 2.5 - FTP Directory Listing
Windows QTL Cartographer 2.5 - FTP Directory Listing
Windows QTL Cartographer 2.5 - FTP Directory Listing
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
52<br />
<strong>Windows</strong> <strong>QTL</strong> <strong>Cartographer</strong> <strong>2.5</strong><br />
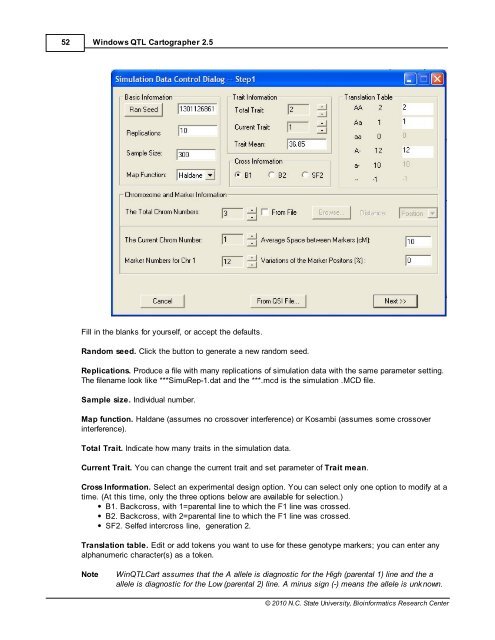
Fill in the blanks for yourself, or accept the defaults.<br />
Random seed. Click the button to generate a new random seed.<br />
Replications. Produce a file with many replications of simulation data with the same parameter setting.<br />
The filename look like ***SimuRep-1.dat and the ***.mcd is the simulation .MCD file.<br />
Sample size. Individual number.<br />
Map function. Haldane (assumes no crossover interference) or Kosambi (assumes some crossover<br />
interference).<br />
Total Trait. Indicate how many traits in the simulation data.<br />
Current Trait. You can change the current trait and set parameter of Trait mean.<br />
Cross Information. Select an experimental design option. You can select only one option to modify at a<br />
time. (At this time, only the three options below are available for selection.)<br />
B1. Backcross, with 1=parental line to which the F1 line was crossed.<br />
B2. Backcross, with 2=parental line to which the F1 line was crossed.<br />
SF2. Selfed intercross line, generation 2.<br />
Translation table. Edit or add tokens you want to use for these genotype markers; you can enter any<br />
alphanumeric character(s) as a token.<br />
Note Win<strong>QTL</strong>Cart assumes that the A allele is diagnostic for the High (parental 1) line and the a<br />
allele is diagnostic for the Low (parental 2) line. A minus sign (-) means the allele is unknown.<br />
© 2010 N.C. State University, Bioinformatics Research Center