Windows QTL Cartographer 2.5 - FTP Directory Listing
Windows QTL Cartographer 2.5 - FTP Directory Listing
Windows QTL Cartographer 2.5 - FTP Directory Listing
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
© 2010 N.C. State University, Bioinformatics Research Center<br />
Win<strong>QTL</strong>Cart Procedures 57<br />
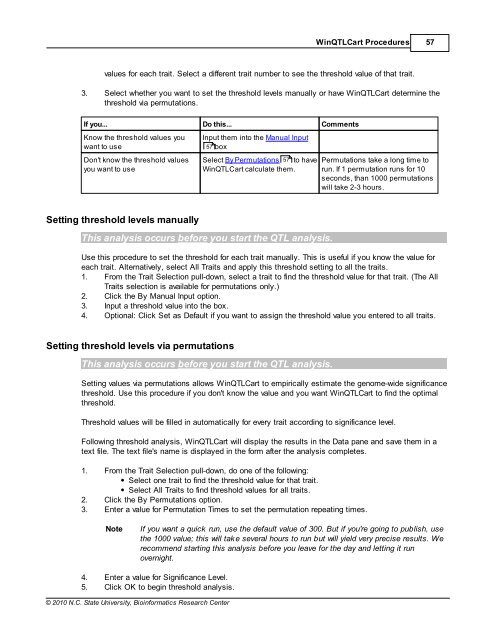
values for each trait. Select a different trait number to see the threshold value of that trait.<br />
3. Select whether you want to set the threshold levels manually or have Win<strong>QTL</strong>Cart determine the<br />
threshold via permutations.<br />
If you... Do this... Comments<br />
Know the threshold values you<br />
want to use<br />
Don't know the threshold values<br />
you want to use<br />
Setting threshold levels manually<br />
Input them into the Manual Input<br />
57 box<br />
Select By Permutations 57<br />
to have<br />
Win<strong>QTL</strong>Cart calculate them.<br />
This analysis occurs before you start the <strong>QTL</strong> analysis.<br />
Permutations take a long time to<br />
run. If 1 permutation runs for 10<br />
seconds, than 1000 permutations<br />
will take 2-3 hours.<br />
Use this procedure to set the threshold for each trait manually. This is useful if you know the value for<br />
each trait. Alternatively, select All Traits and apply this threshold setting to all the traits.<br />
1. From the Trait Selection pull-down, select a trait to find the threshold value for that trait. (The All<br />
Traits selection is available for permutations only.)<br />
2. Click the By Manual Input option.<br />
3. Input a threshold value into the box.<br />
4. Optional: Click Set as Default if you want to assign the threshold value you entered to all traits.<br />
Setting threshold levels via permutations<br />
This analysis occurs before you start the <strong>QTL</strong> analysis.<br />
Setting values via permutations allows Win<strong>QTL</strong>Cart to empirically estimate the genome-wide significance<br />
threshold. Use this procedure if you don't know the value and you want Win<strong>QTL</strong>Cart to find the optimal<br />
threshold.<br />
Threshold values will be filled in automatically for every trait according to significance level.<br />
Following threshold analysis, Win<strong>QTL</strong>Cart will display the results in the Data pane and save them in a<br />
text file. The text file's name is displayed in the form after the analysis completes.<br />
1. From the Trait Selection pull-down, do one of the following:<br />
Select one trait to find the threshold value for that trait.<br />
Select All Traits to find threshold values for all traits.<br />
2. Click the By Permutations option.<br />
3. Enter a value for Permutation Times to set the permutation repeating times.<br />
Note If you want a quick run, use the default value of 300. But if you're going to publish, use<br />
the 1000 value; this will take several hours to run but will yield very precise results. We<br />
recommend starting this analysis before you leave for the day and letting it run<br />
overnight.<br />
4. Enter a value for Significance Level.<br />
5. Click OK to begin threshold analysis.