198 Topics in Current Chemistry Editorial Board: A. de Meijere KN ...
198 Topics in Current Chemistry Editorial Board: A. de Meijere KN ...
198 Topics in Current Chemistry Editorial Board: A. de Meijere KN ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Directional Aspects of Intermolecular Interactions 51<br />
suitable for b<strong>in</strong>d<strong>in</strong>g to a nucleic acid. In each case these are protuberances from<br />
a fairly spherical prote<strong>in</strong> that can then <strong>in</strong>teract with one of the grooves of DNA.<br />
Specific <strong>in</strong>teractions of these types allow the prote<strong>in</strong> to regulate gene expression.<br />
The recognition of the bases <strong>in</strong> DNA by hydrogen bond<strong>in</strong>g to the prote<strong>in</strong> <strong>in</strong>volves<br />
patterns, shown <strong>in</strong> Fig. 39. Each comb<strong>in</strong>ation of base pairs <strong>in</strong> a nucleic<br />
acid – G◊ C, A◊ T, C ◊ G, and T◊ A – can be differentiated <strong>in</strong> the major groove by a<br />
hydrogen-bond pattern [103]. The recognition is less specific <strong>in</strong> the m<strong>in</strong>or<br />
groove. The si<strong>de</strong> cha<strong>in</strong>s of prote<strong>in</strong>s available for such recognition are arg<strong>in</strong><strong>in</strong>e<br />
with two hydrogen-bond donor groups <strong>in</strong> the required orientation, glutamic or<br />
aspartic acid with two hydrogen-bond acceptors, and asparag<strong>in</strong>e and glutam<strong>in</strong>e<br />
which conta<strong>in</strong> one donor and one acceptor.<br />
Many crystal structures that <strong>in</strong>volve prote<strong>in</strong>-nucleic acid <strong>in</strong>teractions have<br />
been published <strong>in</strong> the scientific literature recently. Some show this recognition<br />
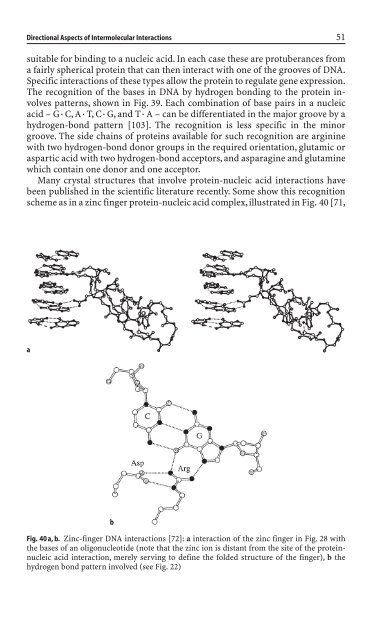
scheme as <strong>in</strong> a z<strong>in</strong>c f<strong>in</strong>ger prote<strong>in</strong>-nucleic acid complex,illustrated <strong>in</strong> Fig. 40 [71,<br />
a<br />
b<br />
Fig. 40 a, b. Z<strong>in</strong>c-f<strong>in</strong>ger DNA <strong>in</strong>teractions [72]: a <strong>in</strong>teraction of the z<strong>in</strong>c f<strong>in</strong>ger <strong>in</strong> Fig. 28 with<br />
the bases of an oligonucleoti<strong>de</strong> (note that the z<strong>in</strong>c ion is distant from the site of the prote<strong>in</strong>nucleic<br />
acid <strong>in</strong>teraction, merely serv<strong>in</strong>g to <strong>de</strong>f<strong>in</strong>e the fol<strong>de</strong>d structure of the f<strong>in</strong>ger), b the<br />
hydrogen bond pattern <strong>in</strong>volved (see Fig. 22)