Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
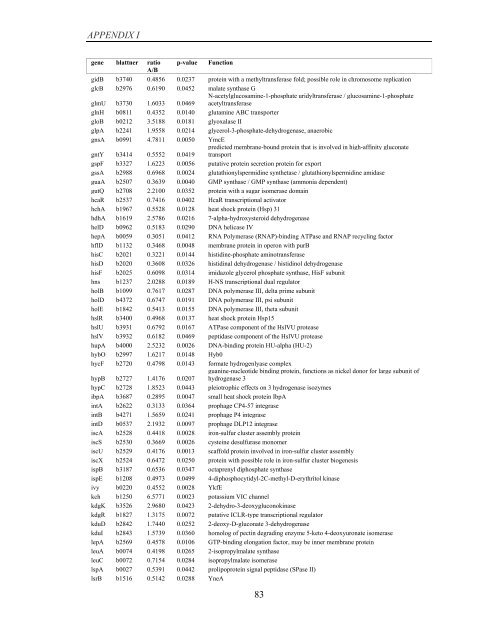
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
gidB b3740 0.4856 0.0237 protein with a methyltransferase fold; possible role in chromosome replication<br />
glcB b2976 0.6190 0.0452 malate synthase G<br />
glmU b3730 1.6033 0.0469<br />
N-acetylglucosamine-1-phosphate uridyltransferase / glucosamine-1-phosphate<br />
acetyltransferase<br />
glnH b0811 0.4352 0.0140 glutamine ABC transporter<br />
gloB b0212 3.5188 0.0181 glyoxalase II<br />
glpA b2241 1.9558 0.0214 glycerol-3-phosphate-dehydrogenase, anaerobic<br />
gnsA b0991 4.7811 0.0050 YmcE<br />
gntY b3414 0.5552 0.0419<br />
predicted membrane-bound protein that is involved in high-affinity gluconate<br />
transport<br />
gspF b3327 1.6223 0.0056 putative protein secretion protein for export<br />
gssA b2988 0.6968 0.0024 glutathionylspermidine synthetase / glutathionylspermidine amidase<br />
guaA b2507 0.3639 0.0040 GMP synthase / GMP synthase (ammonia dependent)<br />
gutQ b2708 2.2100 0.0352 protein with a sugar isomerase domain<br />
hcaR b2537 0.7416 0.0402 HcaR transcriptional activator<br />
hchA b1967 0.5528 0.0128 heat shock protein (Hsp) 31<br />
hdhA b1619 2.5786 0.0216 7-alpha-hydroxysteroid dehydrogenase<br />
helD b0962 0.5183 0.0290 DNA helicase IV<br />
hepA b0059 0.3051 0.0412 RNA Polymerase (RNAP)-binding ATPase and RNAP recycling factor<br />
hflD b1132 0.3468 0.0048 membrane protein in operon with purB<br />
hisC b2021 0.3221 0.0144 histidine-phosphate aminotransferase<br />
hisD b2020 0.3608 0.0326 histidinal dehydrogenase / histidinol dehydrogenase<br />
hisF b2025 0.6098 0.0314 imidazole glycerol phosphate synthase, HisF subunit<br />
hns b1237 2.0288 0.0189 H-NS transcriptional dual regulator<br />
holB b1099 0.7617 0.0287 DNA polymerase III, delta prime subunit<br />
holD b4372 0.6747 0.0191 DNA polymerase III, psi subunit<br />
holE b1842 0.5413 0.0155 DNA polymerase III, theta subunit<br />
hslR b3400 0.4968 0.0137 heat shock protein Hsp15<br />
hslU b3931 0.6792 0.0167 ATPase component <strong>of</strong> the HslVU protease<br />
hslV b3932 0.6182 0.0469 peptidase component <strong>of</strong> the HslVU protease<br />
hupA b4000 2.5232 0.0026 DNA-binding protein HU-alpha (HU-2)<br />
hybO b2997 1.6217 0.0148 Hyb0<br />
hycF b2720 0.4798 0.0143 formate hydrogenlyase complex<br />
hypB b2727 1.4176 0.0207<br />
guanine-nucleotide binding protein, functions as nickel donor for large subunit <strong>of</strong><br />
hydrogenase 3<br />
hypC b2728 1.8523 0.0443 pleiotrophic effects on 3 hydrogenase isozymes<br />
ibpA b3687 0.2895 0.0047 small heat shock protein IbpA<br />
intA b2622 0.3133 0.0364 prophage CP4-57 integrase<br />
intB b4271 1.5659 0.0241 prophage P4 integrase<br />
intD b0537 2.1932 0.0097 prophage DLP12 integrase<br />
iscA b2528 0.4418 0.0028 iron-sulfur cluster assembly protein<br />
iscS b2530 0.3669 0.0026 cysteine desulfurase monomer<br />
iscU b2529 0.4176 0.0013 scaffold protein involved in iron-sulfur cluster assembly<br />
iscX b2524 0.6472 0.0250 protein with possible role in iron-sulfur cluster bio<strong>gene</strong>sis<br />
ispB b3187 0.6536 0.0347 octaprenyl diphosphate synthase<br />
ispE b1208 0.4973 0.0499 4-diphosphocytidyl-2C-methyl-D-erythritol kinase<br />
ivy b0220 0.4552 0.0028 YkfE<br />
kch b1250 6.5771 0.0023 potassium VIC channel<br />
kdgK b3526 2.9680 0.0423 2-dehydro-3-deoxygluconokinase<br />
kdgR b1827 1.3175 0.0072 putative ICLR-type transcriptional regulator<br />
kduD b2842 1.7440 0.0252 2-deoxy-D-gluconate 3-dehydrogenase<br />
kduI b2843 1.5739 0.0360 homolog <strong>of</strong> pectin degrading enzyme 5-keto 4-deoxyuronate isomerase<br />
lepA b2569 0.4578 0.0106 GTP-binding elongation factor, may be inner membrane protein<br />
leuA b0074 0.4198 0.0265 2-isopropylmalate synthase<br />
leuC b0072 0.7154 0.0284 isopropylmalate isomerase<br />
lspA b0027 0.5391 0.0442 prolipoprotein signal peptidase (SPase II)<br />
lsrB b1516 0.5142 0.0288 YneA<br />
83