Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
APPENDIX I<br />
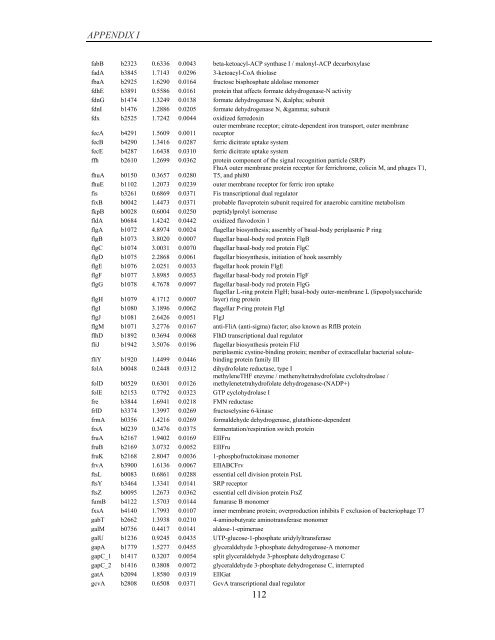
fabB b2323 0.6336 0.0043 beta-ketoacyl-ACP synthase I / malonyl-ACP decarboxylase<br />
fadA b3845 1.7143 0.0296 3-ketoacyl-CoA thiolase<br />
fbaA b2925 1.6290 0.0164 fructose bisphosphate aldolase monomer<br />
fdhE b3891 0.5586 0.0161 protein that affects formate dehydrogenase-N activity<br />
fdnG b1474 1.3249 0.0138 formate dehydrogenase N, α subunit<br />
fdnI b1476 1.2886 0.0205 formate dehydrogenase N, γ subunit<br />
fdx b2525 1.7242 0.0044 oxidized ferredoxin<br />
fecA b4291 1.5609 0.0011<br />
outer membrane receptor; citrate-dependent iron transport, outer membrane<br />
receptor<br />
fecB b4290 1.3416 0.0287 ferric dicitrate uptake system<br />
fecE b4287 1.6438 0.0310 ferric dicitrate uptake system<br />
ffh b2610 1.2699 0.0362 protein component <strong>of</strong> the signal recognition particle (SRP)<br />
fhuA b0150 0.3657 0.0280<br />
FhuA outer membrane protein receptor for ferrichrome, colicin M, and phages T1,<br />
T5, and phi80<br />
fhuE b1102 1.2073 0.0239 outer membrane receptor for ferric iron uptake<br />
fis b3261 0.6869 0.0371 Fis transcriptional dual regulator<br />
fixB b0042 1.4473 0.0371 probable flavoprotein subunit required for anaerobic carnitine metabolism<br />
fkpB b0028 0.6004 0.0250 peptidylprolyl isomerase<br />
fldA b0684 1.4242 0.0442 oxidized flavodoxin 1<br />
flgA b1072 4.8974 0.0024 flagellar biosynthesis; assembly <strong>of</strong> basal-body periplasmic P ring<br />
flgB b1073 3.8020 0.0007 flagellar basal-body rod protein FlgB<br />
flgC b1074 3.0031 0.0070 flagellar basal-body rod protein FlgC<br />
flgD b1075 2.2868 0.0061 flagellar biosynthesis, initiation <strong>of</strong> hook assembly<br />
flgE b1076 2.0251 0.0033 flagellar hook protein FlgE<br />
flgF b1077 3.8985 0.0053 flagellar basal-body rod protein FlgF<br />
flgG b1078 4.7678 0.0097 flagellar basal-body rod protein FlgG<br />
flgH b1079 4.1712 0.0007<br />
flagellar L-ring protein FlgH; basal-body outer-membrane L (lipopolysaccharide<br />
layer) ring protein<br />
flgI b1080 3.1896 0.0062 flagellar P-ring protein FlgI<br />
flgJ b1081 2.6426 0.0051 FlgJ<br />
flgM b1071 3.2776 0.0167 anti-FliA (anti-sigma) factor; also known as RflB protein<br />
flhD b1892 0.3694 0.0068 FlhD transcriptional dual regulator<br />
fliJ b1942 3.5076 0.0196 flagellar biosynthesis protein FliJ<br />
fliY b1920 1.4499 0.0446<br />
periplasmic cystine-binding protein; member <strong>of</strong> extracellular bacterial solutebinding<br />
protein family III<br />
folA b0048 0.2448 0.0312 dihydr<strong>of</strong>olate reductase, type I<br />
folD b0529 0.6301 0.0126<br />
methyleneTHF enzyme / methenyltetrahydr<strong>of</strong>olate cyclohydrolase /<br />
methylenetetrahydr<strong>of</strong>olate dehydrogenase-(NADP+)<br />
folE b2153 0.7792 0.0323 GTP cyclohydrolase I<br />
fre b3844 1.6941 0.0218 FMN reductase<br />
frlD b3374 1.3997 0.0269 fructoselysine 6-kinase<br />
frmA b0356 1.4216 0.0269 formaldehyde dehydrogenase, glutathione-dependent<br />
frsA b0239 0.3476 0.0375 fermentation/respiration switch protein<br />
fruA b2167 1.9402 0.0169 EIIFru<br />
fruB b2169 3.0732 0.0052 EIIFru<br />
fruK b2168 2.8047 0.0036 1-phosph<strong>of</strong>ructokinase monomer<br />
frvA b3900 1.6136 0.0067 EIIABCFrv<br />
ftsL b0083 0.6861 0.0288 essential cell division protein FtsL<br />
ftsY b3464 1.3341 0.0141 SRP receptor<br />
ftsZ b0095 1.2673 0.0362 essential cell division protein FtsZ<br />
fumB b4122 1.5703 0.0144 fumarase B monomer<br />
fxsA b4140 1.7993 0.0107 inner membrane protein; overproduction inhibits F exclusion <strong>of</strong> bacteriophage T7<br />
gabT b2662 1.3938 0.0210 4-aminobutyrate aminotransferase monomer<br />
galM b0756 0.4417 0.0141 aldose-1-epimerase<br />
galU b1236 0.9245 0.0435 UTP-glucose-1-phosphate uridylyltransferase<br />
gapA b1779 1.5277 0.0455 glyceraldehyde 3-phosphate dehydrogenase-A monomer<br />
gapC_1 b1417 0.3207 0.0054 split glyceraldehyde 3-phosphate dehydrogenase C<br />
gapC_2 b1416 0.3808 0.0072 glyceraldehyde 3-phosphate dehydrogenase C, interrupted<br />
gatA b2094 1.8580 0.0319 EIIGat<br />
gcvA b2808 0.6508 0.0371 GcvA transcriptional dual regulator<br />
112