Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
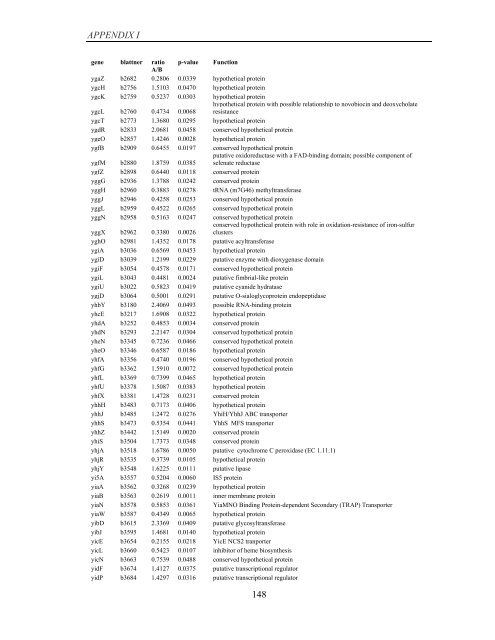
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
ygaZ b2682 0.2806 0.0339 hypothetical protein<br />
ygcH b2756 1.5103 0.0470 hypothetical protein<br />
ygcK b2759 0.5237 0.0303 hypothetical protein<br />
ygcL b2760 0.4734 0.0068<br />
hypothetical protein with possible relationship to novobiocin and deoxycholate<br />
resistance<br />
ygcT b2773 1.3680 0.0295 hypothetical protein<br />
ygdR b2833 2.0681 0.0458 conserved hypothetical protein<br />
ygeO b2857 1.4246 0.0028 hypothetical protein<br />
ygfB b2909 0.6455 0.0197 conserved hypothetical protein<br />
ygfM b2880 1.8759 0.0385<br />
putative oxidoreductase with a FAD-binding domain; possible component <strong>of</strong><br />
selenate reductase<br />
ygfZ b2898 0.6440 0.0118 conserved protein<br />
yggG b2936 1.3788 0.0242 conserved protein<br />
yggH b2960 0.3883 0.0278 tRNA (m7G46) methyltransferase<br />
yggJ b2946 0.4258 0.0253 conserved hypothetical protein<br />
yggL b2959 0.4522 0.0265 conserved hypothetical protein<br />
yggN b2958 0.5163 0.0247 conserved hypothetical protein<br />
yggX b2962 0.3380 0.0026<br />
conserved hypothetical protein with role in oxidation-resistance <strong>of</strong> iron-sulfur<br />
clusters<br />
yghO b2981 1.4352 0.0178 putative acyltransferase<br />
ygiA b3036 0.6569 0.0453 hypothetical protein<br />
ygiD b3039 1.2199 0.0229 putative enzyme with dioxygenase domain<br />
ygiF b3054 0.4578 0.0171 conserved hypothetical protein<br />
ygiL b3043 0.4481 0.0024 putative fimbrial-like protein<br />
ygiU b3022 0.5823 0.0419 putative cyanide hydratase<br />
ygjD b3064 0.5001 0.0291 putative O-sialoglycoprotein endopeptidase<br />
yhbY b3180 2.4069 0.0493 possible RNA-binding protein<br />
yhcE b3217 1.6908 0.0322 hypothetical protein<br />
yhdA b3252 0.4853 0.0034 conserved protein<br />
yhdN b3293 2.2147 0.0304 conserved hypothetical protein<br />
yheN b3345 0.7236 0.0466 conserved hypothetical protein<br />
yheO b3346 0.6587 0.0186 hypothetical protein<br />
yhfA b3356 0.4740 0.0196 conserved hypothetical protein<br />
yhfG b3362 1.5910 0.0072 conserved hypothetical protein<br />
yhfL b3369 0.7399 0.0465 hypothetical protein<br />
yhfU b3378 1.5087 0.0383 hypothetical protein<br />
yhfX b3381 1.4728 0.0231 conserved protein<br />
yhhH b3483 0.7173 0.0406 hypothetical protein<br />
yhhJ b3485 1.2472 0.0276 YhiH/YhhJ ABC transporter<br />
yhhS b3473 0.5354 0.0441 YhhS MFS transporter<br />
yhhZ b3442 1.5149 0.0020 conserved protein<br />
yhiS b3504 1.7373 0.0348 conserved protein<br />
yhjA b3518 1.6786 0.0050 putative cytochrome C peroxidase (EC 1.11.1)<br />
yhjR b3535 0.3739 0.0105 hypothetical protein<br />
yhjY b3548 1.6225 0.0111 putative lipase<br />
yi5A b3557 0.5204 0.0060 IS5 protein<br />
yiaA b3562 0.3268 0.0239 hypothetical protein<br />
yiaB b3563 0.2619 0.0011 inner membrane protein<br />
yiaN b3578 0.5853 0.0361 YiaMNO Binding Protein-dependent Secondary (TRAP) Transporter<br />
yiaW b3587 0.4349 0.0065 hypothetical protein<br />
yibD b3615 2.3369 0.0409 putative glycosyltransferase<br />
yibJ b3595 1.4681 0.0140 hypothetical protein<br />
yicE b3654 0.2155 0.0218 YicE NCS2 tranporter<br />
yicL b3660 0.5423 0.0107 inhibitor <strong>of</strong> heme biosynthesis<br />
yicN b3663 0.7539 0.0488 conserved hypothetical protein<br />
yidF b3674 1.4127 0.0375 putative transcriptional regulator<br />
yidP b3684 1.4297 0.0316 putative transcriptional regulator<br />
148