Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
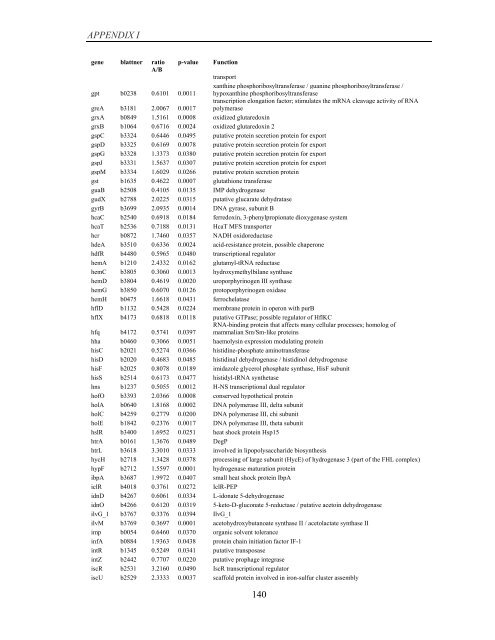
APPENDIX I<br />
<strong>gene</strong> blattner ratio<br />
A/B<br />
p-value Function<br />
transport<br />
gpt b0238 0.6101 0.0011<br />
xanthine phosphoribosyltransferase / guanine phosphoribosyltransferase /<br />
hypoxanthine phosphoribosyltransferase<br />
greA b3181 2.0067 0.0017<br />
transcription elongation factor; stimulates the mRNA cleavage activity <strong>of</strong> RNA<br />
polymerase<br />
grxA b0849 1.5161 0.0008 oxidized glutaredoxin<br />
grxB b1064 0.6716 0.0024 oxidized glutaredoxin 2<br />
gspC b3324 0.6446 0.0495 putative protein secretion protein for export<br />
gspD b3325 0.6169 0.0078 putative protein secretion protein for export<br />
gspG b3328 1.3373 0.0380 putative protein secretion protein for export<br />
gspJ b3331 1.5637 0.0307 putative protein secretion protein for export<br />
gspM b3334 1.6029 0.0266 putative protein secretion protein<br />
gst b1635 0.4622 0.0007 glutathione transferase<br />
guaB b2508 0.4105 0.0135 IMP dehydrogenase<br />
gudX b2788 2.0225 0.0315 putative glucarate dehydratase<br />
gyrB b3699 2.0935 0.0014 DNA gyrase, subunit B<br />
hcaC b2540 0.6918 0.0184 ferredoxin, 3-phenylpropionate dioxygenase system<br />
hcaT b2536 0.7188 0.0131 HcaT MFS transporter<br />
hcr b0872 1.7460 0.0357 NADH oxidoreductase<br />
hdeA b3510 0.6336 0.0024 acid-resistance protein, possible chaperone<br />
hdfR b4480 0.5965 0.0480 transcriptional regulator<br />
hemA b1210 2.4332 0.0162 glutamyl-tRNA reductase<br />
hemC b3805 0.3060 0.0013 hydroxymethylbilane synthase<br />
hemD b3804 0.4619 0.0020 uroporphyrinogen III synthase<br />
hemG b3850 0.6070 0.0126 protoporphyrinogen oxidase<br />
hemH b0475 1.6618 0.0431 ferrochelatase<br />
hflD b1132 0.5428 0.0224 membrane protein in operon with purB<br />
hflX b4173 0.6818 0.0118 putative GTPase; possible regulator <strong>of</strong> HflKC<br />
hfq b4172 0.5741 0.0397<br />
RNA-binding protein that affects many cellular processes; homolog <strong>of</strong><br />
mammalian Sm/Sm-like proteins<br />
hha b0460 0.3066 0.0051 haemolysin <strong>expression</strong> modulating protein<br />
hisC b2021 0.5274 0.0366 histidine-phosphate aminotransferase<br />
hisD b2020 0.4683 0.0485 histidinal dehydrogenase / histidinol dehydrogenase<br />
hisF b2025 0.8078 0.0189 imidazole glycerol phosphate synthase, HisF subunit<br />
hisS b2514 0.6173 0.0477 histidyl-tRNA synthetase<br />
hns b1237 0.5055 0.0012 H-NS transcriptional dual regulator<br />
h<strong>of</strong>O b3393 2.0366 0.0008 conserved hypothetical protein<br />
holA b0640 1.8168 0.0002 DNA polymerase III, delta subunit<br />
holC b4259 0.2779 0.0200 DNA polymerase III, chi subunit<br />
holE b1842 0.2376 0.0017 DNA polymerase III, theta subunit<br />
hslR b3400 1.6952 0.0251 heat shock protein Hsp15<br />
htrA b0161 1.3676 0.0489 DegP<br />
htrL b3618 3.3010 0.0333 involved in lipopolysaccharide biosynthesis<br />
hycH b2718 1.3428 0.0378 processing <strong>of</strong> large subunit (HycE) <strong>of</strong> hydrogenase 3 (part <strong>of</strong> the FHL complex)<br />
hypF b2712 1.5597 0.0001 hydrogenase maturation protein<br />
ibpA b3687 1.9972 0.0407 small heat shock protein IbpA<br />
iclR b4018 0.3761 0.0272 IclR-PEP<br />
idnD b4267 0.6061 0.0334 L-idonate 5-dehydrogenase<br />
idnO b4266 0.6120 0.0319 5-keto-D-gluconate 5-reductase / putative acetoin dehydrogenase<br />
ilvG_1 b3767 0.3376 0.0394 IlvG_1<br />
ilvM b3769 0.3697 0.0001 acetohydroxybutanoate synthase II / acetolactate synthase II<br />
imp b0054 0.6460 0.0370 organic solvent tolerance<br />
infA b0884 1.9363 0.0438 protein chain initiation factor IF-1<br />
intR b1345 0.5249 0.0341 putative transposase<br />
intZ b2442 0.7707 0.0220 putative prophage integrase<br />
iscR b2531 3.2160 0.0490 IscR transcriptional regulator<br />
iscU b2529 2.3333 0.0037 scaffold protein involved in iron-sulfur cluster assembly<br />
140