Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
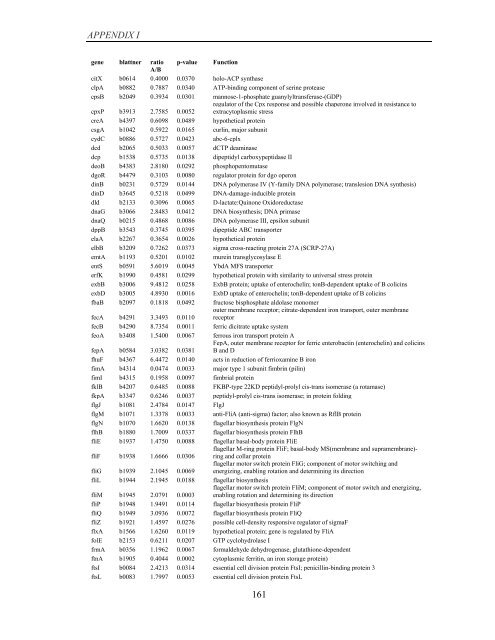
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
citX b0614 0.4000 0.0370 holo-ACP synthase<br />
clpA b0882 0.7887 0.0340 ATP-binding component <strong>of</strong> serine protease<br />
cpsB b2049 0.3934 0.0301 mannose-1-phosphate guanylyltransferase-(GDP)<br />
cpxP b3913 2.7585 0.0052<br />
regulator <strong>of</strong> the Cpx response and possible chaperone involved in resistance to<br />
extracytoplasmic stress<br />
creA b4397 0.6098 0.0489 hypothetical protein<br />
csgA b1042 0.5922 0.0165 curlin, major subunit<br />
cydC b0886 0.5727 0.0423 abc-6-cplx<br />
dcd b2065 0.5033 0.0057 dCTP deaminase<br />
dcp b1538 0.5735 0.0138 dipeptidyl carboxypeptidase II<br />
deoB b4383 2.8180 0.0292 phosphopentomutase<br />
dgoR b4479 0.3103 0.0080 regulator protein for dgo operon<br />
dinB b0231 0.5729 0.0144 DNA polymerase IV (Y-family DNA polymerase; translesion DNA synthesis)<br />
dinD b3645 0.5218 0.0499 DNA-damage-inducible protein<br />
dld b2133 0.3096 0.0065 D-lactate:Quinone Oxidoreductase<br />
dnaG b3066 2.8483 0.0412 DNA biosynthesis; DNA primase<br />
dnaQ b0215 0.4868 0.0086 DNA polymerase III, epsilon subunit<br />
dppB b3543 0.3745 0.0395 dipeptide ABC transporter<br />
elaA b2267 0.3654 0.0026 hypothetical protein<br />
elbB b3209 0.7262 0.0373 sigma cross-reacting protein 27A (SCRP-27A)<br />
emtA b1193 0.5201 0.0102 murein transglycosylase E<br />
entS b0591 5.6019 0.0045 YbdA MFS transporter<br />
erfK b1990 0.4581 0.0299 hypothetical protein with similarity to universal stress protein<br />
exbB b3006 9.4812 0.0258 ExbB protein; uptake <strong>of</strong> enterochelin; tonB-dependent uptake <strong>of</strong> B colicins<br />
exbD b3005 4.8930 0.0016 ExbD uptake <strong>of</strong> enterochelin; tonB-dependent uptake <strong>of</strong> B colicins<br />
fbaB b2097 0.1818 0.0492 fructose bisphosphate aldolase monomer<br />
fecA b4291 3.3493 0.0110<br />
outer membrane receptor; citrate-dependent iron transport, outer membrane<br />
receptor<br />
fecB b4290 8.7354 0.0011 ferric dicitrate uptake system<br />
feoA b3408 1.5400 0.0067 ferrous iron transport protein A<br />
fepA b0584 3.0382 0.0381<br />
FepA, outer membrane receptor for ferric enterobactin (enterochelin) and colicins<br />
B and D<br />
fhuF b4367 6.4472 0.0140 acts in reduction <strong>of</strong> ferrioxamine B iron<br />
fimA b4314 0.0474 0.0033 major type 1 subunit fimbrin (pilin)<br />
fimI b4315 0.1958 0.0097 fimbrial protein<br />
fklB b4207 0.6485 0.0088 FKBP-type 22KD peptidyl-prolyl cis-trans isomerase (a rotamase)<br />
fkpA b3347 0.6246 0.0037 peptidyl-prolyl cis-trans isomerase; in protein folding<br />
flgJ b1081 2.4784 0.0147 FlgJ<br />
flgM b1071 1.3378 0.0033 anti-FliA (anti-sigma) factor; also known as RflB protein<br />
flgN b1070 1.6620 0.0138 flagellar biosynthesis protein FlgN<br />
flhB b1880 1.7009 0.0337 flagellar biosynthesis protein FlhB<br />
fliE b1937 1.4750 0.0088 flagellar basal-body protein FliE<br />
fliF b1938 1.6666 0.0306<br />
flagellar M-ring protein FliF; basal-body MS(membrane and supramembrane)-<br />
ring and collar protein<br />
fliG b1939 2.1045 0.0069<br />
flagellar motor switch protein FliG; component <strong>of</strong> motor switching and<br />
energizing, enabling rotation and determining its direction<br />
fliL b1944 2.1945 0.0188 flagellar biosynthesis<br />
fliM b1945 2.0791 0.0003<br />
flagellar motor switch protein FliM; component <strong>of</strong> motor switch and energizing,<br />
enabling rotation and determining its direction<br />
fliP b1948 1.9491 0.0114 flagellar biosynthesis protein FliP<br />
fliQ b1949 3.0936 0.0072 flagellar biosynthesis protein FliQ<br />
fliZ b1921 1.4597 0.0276 possible cell-density responsive regulator <strong>of</strong> sigmaF<br />
flxA b1566 1.6260 0.0119 hypothetical protein; <strong>gene</strong> is regulated <strong>by</strong> FliA<br />
folE b2153 0.6211 0.0207 GTP cyclohydrolase I<br />
frmA b0356 1.1962 0.0067 formaldehyde dehydrogenase, glutathione-dependent<br />
ftnA b1905 0.4044 0.0002 cytoplasmic ferritin, an iron storage protein)<br />
ftsI b0084 2.4213 0.0314 essential cell division protein FtsI; penicillin-binding protein 3<br />
ftsL b0083 1.7997 0.0053 essential cell division protein FtsL<br />
161