Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
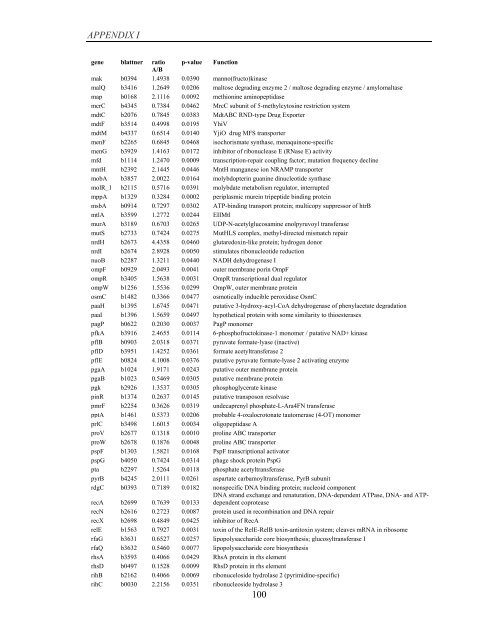
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
mak b0394 1.4938 0.0390 manno(fructo)kinase<br />
malQ b3416 1.2649 0.0206 maltose degrading enzyme 2 / maltose degrading enzyme / amylomaltase<br />
map b0168 2.1116 0.0092 methionine aminopeptidase<br />
mcrC b4345 0.7384 0.0462 MrcC subunit <strong>of</strong> 5-methylcytosine restriction system<br />
mdtC b2076 0.7845 0.0383 MdtABC RND-type Drug Exporter<br />
mdtF b3514 0.4998 0.0195 YhiV<br />
mdtM b4337 0.6514 0.0140 YjiO drug MFS transporter<br />
menF b2265 0.6845 0.0468 isochorismate synthase, menaquinone-specific<br />
menG b3929 1.4163 0.0172 inhibitor <strong>of</strong> ribonuclease E (RNase E) activity<br />
mfd b1114 1.2470 0.0009 transcription-repair coupling factor; mutation frequency decline<br />
mntH b2392 2.1445 0.0446 MntH manganese ion NRAMP transporter<br />
mobA b3857 2.0022 0.0164 molybdopterin guanine dinucleotide synthase<br />
molR_1 b2115 0.5716 0.0391 molybdate metabolism regulator, interrupted<br />
mppA b1329 0.3284 0.0002 periplasmic murein tripeptide binding protein<br />
msbA b0914 0.7297 0.0302 ATP-binding transport protein; multicopy suppressor <strong>of</strong> htrB<br />
mtlA b3599 1.2772 0.0244 EIIMtl<br />
murA b3189 0.6703 0.0265 UDP-N-acetylglucosamine enolpyruvoyl transferase<br />
mutS b2733 0.7424 0.0275 MutHLS complex, methyl-directed mismatch repair<br />
nrdH b2673 4.4358 0.0460 glutaredoxin-like protein; hydrogen donor<br />
nrdI b2674 2.8928 0.0050 stimulates ribonucleotide reduction<br />
nuoB b2287 1.3211 0.0440 NADH dehydrogenase I<br />
ompF b0929 2.0493 0.0041 outer membrane porin OmpF<br />
ompR b3405 1.5638 0.0031 OmpR transcriptional dual regulator<br />
ompW b1256 1.5536 0.0299 OmpW, outer membrane protein<br />
osmC b1482 0.3366 0.0477 osmotically inducible peroxidase OsmC<br />
paaH b1395 1.6745 0.0471 putative 3-hydroxy-acyl-CoA dehydrogenase <strong>of</strong> phenylacetate degradation<br />
paaI b1396 1.5659 0.0497 hypothetical protein with some similarity to thioesterases<br />
pagP b0622 0.2030 0.0037 PagP monomer<br />
pfkA b3916 2.4655 0.0114 6-phosph<strong>of</strong>ructokinase-1 monomer / putative NAD+ kinase<br />
pflB b0903 2.0318 0.0371 pyruvate formate-lyase (inactive)<br />
pflD b3951 1.4252 0.0361 formate acetyltransferase 2<br />
pflE b0824 4.1008 0.0376 putative pyruvate formate-lyase 2 activating enzyme<br />
pgaA b1024 1.9171 0.0243 putative outer membrane protein<br />
pgaB b1023 0.5469 0.0305 putative membrane protein<br />
pgk b2926 1.3537 0.0305 phosphoglycerate kinase<br />
pinR b1374 0.2637 0.0145 putative transposon resolvase<br />
pmrF b2254 0.3626 0.0319 undecaprenyl phosphate-L-Ara4FN transferase<br />
pptA b1461 0.5373 0.0206 probable 4-oxalocrotonate tautomerase (4-OT) monomer<br />
prlC b3498 1.6015 0.0034 oligopeptidase A<br />
proV b2677 0.1318 0.0010 proline ABC transporter<br />
proW b2678 0.1876 0.0048 proline ABC transporter<br />
pspF b1303 1.5821 0.0168 PspF transcriptional activator<br />
pspG b4050 0.7424 0.0314 phage shock protein PspG<br />
pta b2297 1.5264 0.0118 phosphate acetyltransferase<br />
pyrB b4245 2.0111 0.0261 aspartate carbamoyltransferase, PyrB subunit<br />
rdgC b0393 0.7189 0.0182 nonspecific DNA binding protein; nucleoid component<br />
recA b2699 0.7639 0.0133<br />
DNA strand exchange and renaturation, DNA-dependent ATPase, DNA- and ATPdependent<br />
coprotease<br />
recN b2616 0.2723 0.0087 protein used in recombination and DNA repair<br />
recX b2698 0.4849 0.0425 inhibitor <strong>of</strong> RecA<br />
relE b1563 0.7927 0.0031 toxin <strong>of</strong> the RelE-RelB toxin-antitoxin system; cleaves mRNA in ribosome<br />
rfaG b3631 0.6527 0.0257 lipopolysaccharide core biosynthesis; glucosyltransferase I<br />
rfaQ b3632 0.5460 0.0077 lipopolysaccharide core biosynthesis<br />
rhsA b3593 0.4066 0.0429 RhsA protein in rhs element<br />
rhsD b0497 0.1528 0.0099 RhsD protein in rhs element<br />
rihB b2162 0.4066 0.0069 ribonuceloside hydrolase 2 (pyrimidine-specific)<br />
rihC b0030 2.2156 0.0351 ribonucleoside hydrolase 3<br />
100