Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
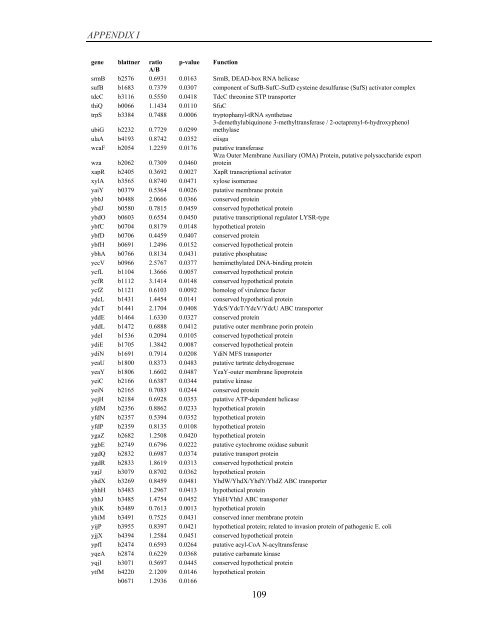
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
srmB b2576 0.6931 0.0163 SrmB, DEAD-box RNA helicase<br />
sufB b1683 0.7379 0.0307 component <strong>of</strong> SufB-SufC-SufD cysteine desulfurase (SufS) activator complex<br />
tdcC b3116 0.5550 0.0418 TdcC threonine STP transporter<br />
thiQ b0066 1.1434 0.0110 SfuC<br />
trpS b3384 0.7488 0.0006 tryptophanyl-tRNA synthetase<br />
ubiG b2232 0.7729 0.0299<br />
3-demethylubiquinone 3-methyltransferase / 2-octaprenyl-6-hydroxyphenol<br />
methylase<br />
ulaA b4193 0.8742 0.0352 eiisga<br />
wcaF b2054 1.2259 0.0176 putative transferase<br />
wza b2062 0.7309 0.0460<br />
Wza Outer Membrane Auxiliary (OMA) Protein, putative polysaccharide export<br />
protein<br />
xapR b2405 0.3692 0.0027 XapR transcriptional activator<br />
xylA b3565 0.8740 0.0471 xylose isomerase<br />
yaiY b0379 0.5364 0.0026 putative membrane protein<br />
ybbJ b0488 2.0666 0.0366 conserved protein<br />
ybdJ b0580 0.7815 0.0459 conserved hypothetical protein<br />
ybdO b0603 0.6554 0.0450 putative transcriptional regulator LYSR-type<br />
ybfC b0704 0.8179 0.0148 hypothetical protein<br />
ybfD b0706 0.4459 0.0407 conserved protein<br />
ybfH b0691 1.2496 0.0152 conserved hypothetical protein<br />
ybhA b0766 0.8134 0.0431 putative phosphatase<br />
yccV b0966 2.5767 0.0377 hemimethylated DNA-binding protein<br />
ycfL b1104 1.3666 0.0057 conserved hypothetical protein<br />
ycfR b1112 3.1414 0.0148 conserved hypothetical protein<br />
ycfZ b1121 0.6103 0.0092 homolog <strong>of</strong> virulence factor<br />
ydcL b1431 1.4454 0.0141 conserved hypothetical protein<br />
ydcT b1441 2.1704 0.0408 YdcS/YdcT/YdcV/YdcU ABC transporter<br />
yddE b1464 1.6330 0.0327 conserved protein<br />
yddL b1472 0.6888 0.0412 putative outer membrane porin protein<br />
ydeI b1536 0.2094 0.0105 conserved hypothetical protein<br />
ydiE b1705 1.3842 0.0087 conserved hypothetical protein<br />
ydiN b1691 0.7914 0.0208 YdiN MFS transporter<br />
yeaU b1800 0.8373 0.0483 putative tartrate dehydrogenase<br />
yeaY b1806 1.6602 0.0487 YeaY-outer membrane lipoprotein<br />
yeiC b2166 0.6387 0.0344 putative kinase<br />
yeiN b2165 0.7083 0.0244 conserved protein<br />
yejH b2184 0.6928 0.0353 putative ATP-dependent helicase<br />
yfdM b2356 0.8862 0.0233 hypothetical protein<br />
yfdN b2357 0.5394 0.0352 hypothetical protein<br />
yfdP b2359 0.8135 0.0108 hypothetical protein<br />
ygaZ b2682 1.2508 0.0420 hypothetical protein<br />
ygbE b2749 0.6796 0.0222 putative cytochrome oxidase subunit<br />
ygdQ b2832 0.6987 0.0374 putative transport protein<br />
ygdR b2833 1.8619 0.0313 conserved hypothetical protein<br />
ygjJ b3079 0.8702 0.0362 hypothetical protein<br />
yhdX b3269 0.8459 0.0481 YhdW/YhdX/YhdY/YhdZ ABC transporter<br />
yhhH b3483 1.2967 0.0413 hypothetical protein<br />
yhhJ b3485 1.4754 0.0452 YhiH/YhhJ ABC transporter<br />
yhiK b3489 0.7613 0.0013 hypothetical protein<br />
yhiM b3491 0.7525 0.0431 conserved inner membrane protein<br />
yijP b3955 0.8397 0.0421 hypothetical protein; related to invasion protein <strong>of</strong> pathogenic E. coli<br />
yjjX b4394 1.2584 0.0451 conserved hypothetical protein<br />
ypfI b2474 0.6593 0.0264 putative acyl-CoA N-acyltransferase<br />
yqeA b2874 0.6229 0.0368 putative carbamate kinase<br />
yqjI b3071 0.5697 0.0445 conserved hypothetical protein<br />
ytfM b4220 2.1209 0.0146 hypothetical protein<br />
b0671 1.2936 0.0166<br />
109