Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
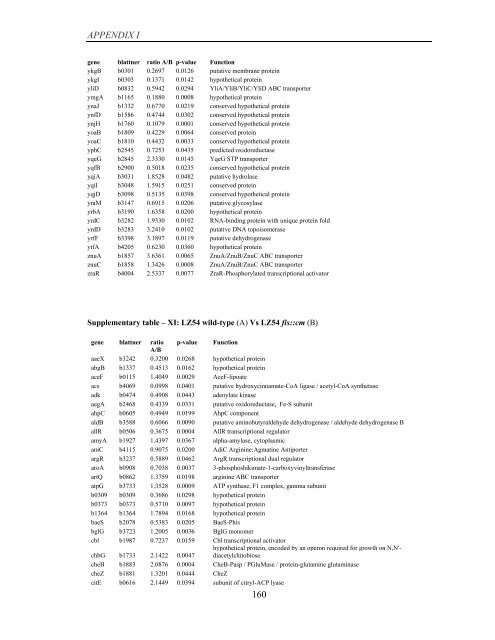
APPENDIX I<br />
<strong>gene</strong> blattner ratio A/B p-value Function<br />
ykgB b0301 0.2697 0.0126 putative membrane protein<br />
ykgI b0303 0.1371 0.0142 hypothetical protein<br />
yliD b0832 0.5942 0.0294 YliA/YliB/YliC/YliD ABC transporter<br />
ymgA b1165 0.1880 0.0008 hypothetical protein<br />
ynaJ b1332 0.6770 0.0219 conserved hypothetical protein<br />
ynfD b1586 0.4744 0.0302 conserved hypothetical protein<br />
ynjH b1760 0.1079 0.0001 conserved hypothetical protein<br />
yoaB b1809 0.4229 0.0064 conserved protein<br />
yoaC b1810 0.4432 0.0033 conserved hypothetical protein<br />
yphC b2545 0.7253 0.0435 predicted oxidoreductase<br />
yqeG b2845 2.3330 0.0145 YqeG STP transporter<br />
yqfB b2900 0.5018 0.0235 conserved hypothetical protein<br />
yqiA b3031 1.8528 0.0482 putative hydrolase<br />
yqiI b3048 1.5915 0.0251 conserved protein<br />
yqjD b3098 0.5135 0.0398 conserved hypothetical protein<br />
yraM b3147 0.6915 0.0206 putative glycosylase<br />
yrbA b3190 1.6358 0.0200 hypothetical protein<br />
yrdC b3282 1.9330 0.0102 RNA-binding protein with unique protein fold<br />
yrdD b3283 3.2410 0.0102 putative DNA topoisomerase<br />
yrfF b3398 3.1897 0.0119 putative dehydrogenase<br />
ytfA b4205 0.6230 0.0360 hypothetical protein<br />
znuA b1857 3.6361 0.0065 ZnuA/ZnuB/ZnuC ABC transporter<br />
znuC b1858 1.3426 0.0008 ZnuA/ZnuB/ZnuC ABC transporter<br />
zraR b4004 2.5337 0.0077 ZraR-Phosphorylated transcriptional activator<br />
Supplementary table – XI: LZ54 wild-type (A) Vs LZ54 fis::cm (B)<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
aaeX b3242 0.3200 0.0268 hypothetical protein<br />
abgB b1337 0.4513 0.0162 hypothetical protein<br />
aceF b0115 1.4049 0.0029 AceF-lipoate<br />
acs b4069 0.0998 0.0401 putative hydroxycinnamate-CoA ligase / acetyl-CoA synthetase<br />
adk b0474 0.4908 0.0443 adenylate kinase<br />
aegA b2468 0.4339 0.0331 putative oxidoreductase, Fe-S subunit<br />
ahpC b0605 0.4949 0.0199 AhpC component<br />
aldB b3588 0.6066 0.0090 putative aminobutyraldehyde dehydrogenase / aldehyde dehydrogenase B<br />
allR b0506 0.3675 0.0004 AllR transcriptional regulator<br />
amyA b1927 1.4397 0.0367 alpha-amylase, cytoplasmic<br />
aniC b4115 0.9075 0.0200 AdiC Arginine:Agmatine Antiporter<br />
argR b3237 0.5889 0.0462 ArgR transcriptional dual regulator<br />
aroA b0908 0.7038 0.0037 3-phosphoshikimate-1-carboxyvinyltransferase<br />
artQ b0862 1.3759 0.0198 arginine ABC transporter<br />
atpG b3733 1.3528 0.0009 ATP synthase, F1 complex, gamma subunit<br />
b0309 b0309 0.3686 0.0298 hypothetical protein<br />
b0373 b0373 0.5710 0.0097 hypothetical protein<br />
b1364 b1364 1.7894 0.0168 hypothetical protein<br />
baeS b2078 0.5383 0.0205 BaeS-Phis<br />
bglG b3723 1.2005 0.0036 BglG monomer<br />
cbl b1987 0.7237 0.0159 Cbl transcriptional activator<br />
chbG b1733 2.1422 0.0047<br />
hypothetical protein, encoded <strong>by</strong> an operon required for growth on N,N'-<br />
diacetylchitobiose<br />
cheB b1883 2.0876 0.0004 CheB-Pasp / PGluMase / protein-glutamine glutaminase<br />
cheZ b1881 1.3201 0.0444 CheZ<br />
citE b0616 2.1449 0.0394 subunit <strong>of</strong> citryl-ACP lyase<br />
160