Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
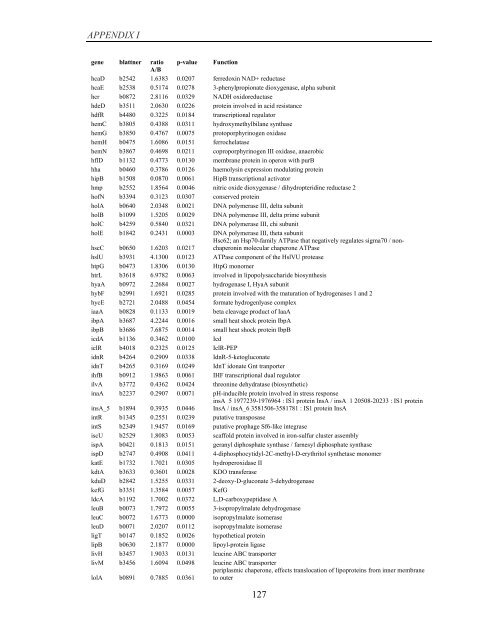
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
hcaD b2542 1.6383 0.0207 ferredoxin NAD+ reductase<br />
hcaE b2538 0.5174 0.0278 3-phenylpropionate dioxygenase, alpha subunit<br />
hcr b0872 2.8116 0.0329 NADH oxidoreductase<br />
hdeD b3511 2.0630 0.0226 protein involved in acid resistance<br />
hdfR b4480 0.3225 0.0184 transcriptional regulator<br />
hemC b3805 0.4388 0.0311 hydroxymethylbilane synthase<br />
hemG b3850 0.4767 0.0075 protoporphyrinogen oxidase<br />
hemH b0475 1.6086 0.0151 ferrochelatase<br />
hemN b3867 0.4698 0.0211 coproporphyrinogen III oxidase, anaerobic<br />
hflD b1132 0.4773 0.0130 membrane protein in operon with purB<br />
hha b0460 0.3786 0.0126 haemolysin <strong>expression</strong> modulating protein<br />
hipB b1508 0.0870 0.0061 HipB transcriptional activator<br />
hmp b2552 1.8564 0.0046 nitric oxide dioxygenase / dihydropteridine reductase 2<br />
h<strong>of</strong>N b3394 0.3123 0.0307 conserved protein<br />
holA b0640 2.0348 0.0021 DNA polymerase III, delta subunit<br />
holB b1099 1.5205 0.0029 DNA polymerase III, delta prime subunit<br />
holC b4259 0.5840 0.0321 DNA polymerase III, chi subunit<br />
holE b1842 0.2431 0.0003 DNA polymerase III, theta subunit<br />
hscC b0650 1.6203 0.0217<br />
Hsc62; an Hsp70-family ATPase that negatively regulates sigma70 / nonchaperonin<br />
molecular chaperone ATPase<br />
hslU b3931 4.1300 0.0123 ATPase component <strong>of</strong> the HslVU protease<br />
htpG b0473 1.8306 0.0130 HtpG monomer<br />
htrL b3618 6.9782 0.0063 involved in lipopolysaccharide biosynthesis<br />
hyaA b0972 2.2684 0.0027 hydrogenase I, HyaA subunit<br />
hybF b2991 1.6921 0.0285 protein involved with the maturation <strong>of</strong> hydrogenases 1 and 2<br />
hycE b2721 2.0488 0.0454 formate hydrogenlyase complex<br />
iaaA b0828 0.1133 0.0019 beta cleavage product <strong>of</strong> IaaA<br />
ibpA b3687 4.2244 0.0016 small heat shock protein IbpA<br />
ibpB b3686 7.6875 0.0014 small heat shock protein IbpB<br />
icdA b1136 0.3462 0.0100 Icd<br />
iclR b4018 0.2325 0.0125 IclR-PEP<br />
idnR b4264 0.2909 0.0338 IdnR-5-ketogluconate<br />
idnT b4265 0.3169 0.0249 IdnT idonate Gnt tranporter<br />
ihfB b0912 1.9863 0.0061 IHF transcriptional dual regulator<br />
ilvA b3772 0.4362 0.0424 threonine dehydratase (biosynthetic)<br />
inaA b2237 0.2907 0.0071 pH-inducible protein involved in stress response<br />
insA_5 b1894 0.3935 0.0446<br />
insA_5 1977239-1976964 : IS1 protein InsA / insA_1 20508-20233 : IS1 protein<br />
InsA / insA_6 3581506-3581781 : IS1 protein InsA<br />
intR b1345 0.2551 0.0239 putative transposase<br />
intS b2349 1.9457 0.0169 putative prophage Sf6-like integrase<br />
iscU b2529 1.8083 0.0053 scaffold protein involved in iron-sulfur cluster assembly<br />
ispA b0421 0.1813 0.0151 geranyl diphosphate synthase / farnesyl diphosphate synthase<br />
ispD b2747 0.4908 0.0411 4-diphosphocytidyl-2C-methyl-D-erythritol synthetase monomer<br />
katE b1732 1.7021 0.0305 hydroperoxidase II<br />
kdtA b3633 0.3601 0.0028 KDO transferase<br />
kduD b2842 1.5255 0.0331 2-deoxy-D-gluconate 3-dehydrogenase<br />
kefG b3351 1.3584 0.0057 KefG<br />
ldcA b1192 1.7002 0.0372 L,D-carboxypeptidase A<br />
leuB b0073 1.7972 0.0055 3-isopropylmalate dehydrogenase<br />
leuC b0072 1.6773 0.0000 isopropylmalate isomerase<br />
leuD b0071 2.0207 0.0112 isopropylmalate isomerase<br />
ligT b0147 0.1852 0.0026 hypothetical protein<br />
lipB b0630 2.1877 0.0000 lipoyl-protein ligase<br />
livH b3457 1.9033 0.0131 leucine ABC transporter<br />
livM b3456 1.6094 0.0498 leucine ABC transporter<br />
lolA b0891 0.7885 0.0361<br />
periplasmic chaperone, effects translocation <strong>of</strong> lipoproteins from inner membrane<br />
to outer<br />
127