Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
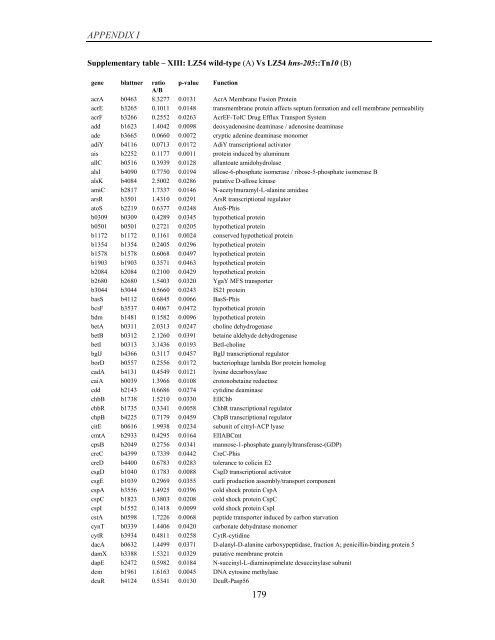
APPENDIX I<br />
Supplementary table – XIII: LZ54 wild-type (A) Vs LZ54 hns-205::Tn10 (B)<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
acrA b0463 8.3277 0.0131 AcrA Membrane Fusion Protein<br />
acrE b3265 0.1011 0.0148 transmembrane protein affects septum formation and cell membrane permeability<br />
acrF b3266 0.2552 0.0263 AcrEF-TolC Drug Efflux Transport System<br />
add b1623 1.4042 0.0098 deoxyadenosine deaminase / adenosine deaminase<br />
ade b3665 0.0660 0.0072 cryptic adenine deaminase monomer<br />
adiY b4116 0.0713 0.0172 AdiY transcriptional activator<br />
ais b2252 0.1177 0.0011 protein induced <strong>by</strong> aluminum<br />
allC b0516 0.3939 0.0128 allantoate amidohydrolase<br />
alsI b4090 0.7750 0.0194 allose-6-phosphate isomerase / ribose-5-phosphate isomerase B<br />
alsK b4084 2.5002 0.0286 putative D-allose kinase<br />
amiC b2817 1.7337 0.0146 N-acetylmuramyl-L-alanine amidase<br />
arsR b3501 1.4310 0.0291 ArsR transcriptional regulator<br />
atoS b2219 0.6377 0.0248 AtoS-Phis<br />
b0309 b0309 0.4289 0.0345 hypothetical protein<br />
b0501 b0501 0.2721 0.0205 hypothetical protein<br />
b1172 b1172 0.1161 0.0024 conserved hypothetical protein<br />
b1354 b1354 0.2405 0.0296 hypothetical protein<br />
b1578 b1578 0.6068 0.0497 hypothetical protein<br />
b1903 b1903 0.3571 0.0463 hypothetical protein<br />
b2084 b2084 0.2100 0.0429 hypothetical protein<br />
b2680 b2680 1.5403 0.0320 YgaY MFS transporter<br />
b3044 b3044 0.5660 0.0243 IS21 protein<br />
basS b4112 0.6845 0.0066 BasS-Phis<br />
bcsF b3537 0.4067 0.0472 hypothetical protein<br />
bdm b1481 0.1582 0.0096 hypothetical protein<br />
betA b0311 2.0313 0.0247 choline dehydrogenase<br />
betB b0312 2.1260 0.0391 betaine aldehyde dehydrogenase<br />
betI b0313 3.1436 0.0193 BetI-choline<br />
bglJ b4366 0.3117 0.0457 BglJ transcriptional regulator<br />
borD b0557 0.2556 0.0172 bacteriophage lambda Bor protein homolog<br />
cadA b4131 0.4549 0.0121 lysine decarboxylase<br />
caiA b0039 1.3966 0.0108 crotonobetaine reductase<br />
cdd b2143 0.6686 0.0274 cytidine deaminase<br />
chbB b1738 1.5210 0.0330 EIIChb<br />
chbR b1735 0.3341 0.0058 ChbR transcriptional regulator<br />
chpB b4225 0.7179 0.0459 ChpB transcriptional regulator<br />
citE b0616 1.9938 0.0234 subunit <strong>of</strong> citryl-ACP lyase<br />
cmtA b2933 0.4295 0.0164 EIIABCmt<br />
cpsB b2049 0.2756 0.0341 mannose-1-phosphate guanylyltransferase-(GDP)<br />
creC b4399 0.7339 0.0442 CreC-Phis<br />
creD b4400 0.6783 0.0283 tolerance to colicin E2<br />
csgD b1040 0.1783 0.0088 CsgD transcriptional activator<br />
csgE b1039 0.2969 0.0355 curli production assembly/transport component<br />
cspA b3556 1.4925 0.0396 cold shock protein CspA<br />
cspC b1823 0.3803 0.0208 cold shock protein CspC<br />
cspI b1552 0.1418 0.0099 cold shock protein CspI<br />
cstA b0598 1.7226 0.0068 peptide transporter induced <strong>by</strong> carbon starvation<br />
cynT b0339 1.4406 0.0420 carbonate dehydratase monomer<br />
cytR b3934 0.4811 0.0258 CytR-cytidine<br />
dacA b0632 1.4499 0.0371 D-alanyl-D-alanine carboxypeptidase, fraction A; penicillin-binding protein 5<br />
damX b3388 1.5321 0.0329 putative membrane protein<br />
dapE b2472 0.5982 0.0184 N-succinyl-L-diaminopimelate desuccinylase subunit<br />
dcm b1961 1.6163 0.0045 DNA cytosine methylase<br />
dcuR b4124 0.5341 0.0130 DcuR-Pasp56<br />
179