Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
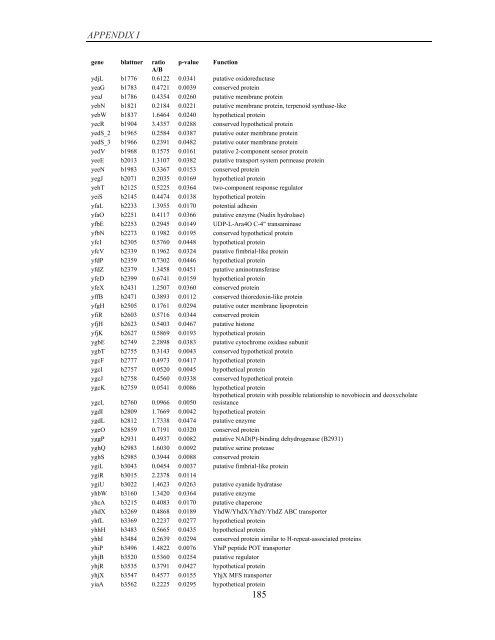
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
ydjL b1776 0.6122 0.0341 putative oxidoreductase<br />
yeaG b1783 0.4721 0.0039 conserved protein<br />
yeaJ b1786 0.4354 0.0260 putative membrane protein<br />
yebN b1821 0.2184 0.0221 putative membrane protein, terpenoid synthase-like<br />
yebW b1837 1.6464 0.0240 hypothetical protein<br />
yecR b1904 3.4357 0.0288 conserved hypothetical protein<br />
yedS_2 b1965 0.2584 0.0387 putative outer membrane protein<br />
yedS_3 b1966 0.2391 0.0482 putative outer membrane protein<br />
yedV b1968 0.1575 0.0161 putative 2-component sensor protein<br />
yeeE b2013 1.3107 0.0382 putative transport system permease protein<br />
yeeN b1983 0.3367 0.0153 conserved protein<br />
yegJ b2071 0.2035 0.0169 hypothetical protein<br />
yehT b2125 0.5225 0.0364 two-component response regulator<br />
yeiS b2145 0.4474 0.0138 hypothetical protein<br />
yfaL b2233 1.3955 0.0170 potential adhesin<br />
yfaO b2251 0.4117 0.0366 putative enzyme (Nudix hydrolase)<br />
yfbE b2253 0.2945 0.0149 UDP-L-Ara4O C-4" transaminase<br />
yfbN b2273 0.1982 0.0195 conserved hypothetical protein<br />
yfcI b2305 0.5760 0.0448 hypothetical protein<br />
yfcV b2339 0.1962 0.0324 putative fimbrial-like protein<br />
yfdP b2359 0.7302 0.0446 hypothetical protein<br />
yfdZ b2379 1.3458 0.0451 putative aminotransferase<br />
yfeD b2399 0.6741 0.0159 hypothetical protein<br />
yfeX b2431 1.2507 0.0360 conserved protein<br />
yffB b2471 0.3893 0.0112 conserved thioredoxin-like protein<br />
yfgH b2505 0.1761 0.0294 putative outer membrane lipoprotein<br />
yfiR b2603 0.5716 0.0344 conserved protein<br />
yfjH b2623 0.5403 0.0467 putative histone<br />
yfjK b2627 0.5869 0.0193 hypothetical protein<br />
ygbE b2749 2.2898 0.0383 putative cytochrome oxidase subunit<br />
ygbT b2755 0.3143 0.0043 conserved hypothetical protein<br />
ygcF b2777 0.4973 0.0417 hypothetical protein<br />
ygcI b2757 0.0520 0.0045 hypothetical protein<br />
ygcJ b2758 0.4560 0.0338 conserved hypothetical protein<br />
ygcK b2759 0.0541 0.0086 hypothetical protein<br />
ygcL b2760 0.0966 0.0050<br />
hypothetical protein with possible relationship to novobiocin and deoxycholate<br />
resistance<br />
ygdI b2809 1.7669 0.0042 hypothetical protein<br />
ygdL b2812 1.7338 0.0474 putative enzyme<br />
ygeO b2859 0.7191 0.0320 conserved protein<br />
yggP b2931 0.4937 0.0082 putative NAD(P)-binding dehydrogenase (B2931)<br />
yghQ b2983 1.6030 0.0092 putative serine protease<br />
yghS b2985 0.3944 0.0088 conserved protein<br />
ygiL b3043 0.0454 0.0037 putative fimbrial-like protein<br />
ygiR b3015 2.2378 0.0114<br />
ygiU b3022 1.4623 0.0263 putative cyanide hydratase<br />
yhbW b3160 1.3420 0.0364 putative enzyme<br />
yhcA b3215 0.4083 0.0170 putative chaperone<br />
yhdX b3269 0.4868 0.0189 YhdW/YhdX/YhdY/YhdZ ABC transporter<br />
yhfL b3369 0.2237 0.0277 hypothetical protein<br />
yhhH b3483 0.5665 0.0435 hypothetical protein<br />
yhhI b3484 0.2639 0.0294 conserved protein similar to H-repeat-associated proteins<br />
yhiP b3496 1.4822 0.0076 YhiP peptide POT transporter<br />
yhjB b3520 0.5360 0.0254 putative regulator<br />
yhjR b3535 0.3791 0.0427 hypothetical protein<br />
yhjX b3547 0.4577 0.0155 YhjX MFS transporter<br />
yiaA b3562 0.2225 0.0295 hypothetical protein<br />
185