Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
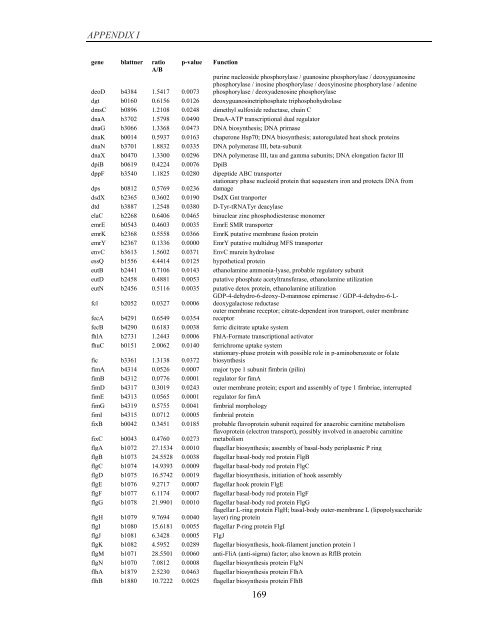
APPENDIX I<br />
<strong>gene</strong> blattner ratio<br />
A/B<br />
p-value<br />
Function<br />
purine nucleoside phosphorylase / guanosine phosphorylase / deoxyguanosine<br />
phosphorylase / inosine phosphorylase / deoxyinosine phosphorylase / adenine<br />
phosphorylase / deoxyadenosine phosphorylase<br />
deoD b4384 1.5417 0.0073<br />
dgt b0160 0.6156 0.0126 deoxyguanosinetriphosphate triphosphohydrolase<br />
dmsC b0896 1.2108 0.0248 dimethyl sulfoxide reductase, chain C<br />
dnaA b3702 1.5798 0.0490 DnaA-ATP transcriptional dual regulator<br />
dnaG b3066 1.3368 0.0473 DNA biosynthesis; DNA primase<br />
dnaK b0014 0.5937 0.0163 chaperone Hsp70; DNA biosynthesis; autoregulated heat shock proteins<br />
dnaN b3701 1.8832 0.0335 DNA polymerase III, beta-subunit<br />
dnaX b0470 1.3300 0.0296 DNA polymerase III, tau and gamma subunits; DNA elongation factor III<br />
dpiB b0619 0.4224 0.0076 DpiB<br />
dppF b3540 1.1825 0.0280 dipeptide ABC transporter<br />
stationary phase nucleoid protein that sequesters iron and protects DNA from<br />
dps b0812 0.5769 0.0236 damage<br />
dsdX b2365 0.3602 0.0190 DsdX Gnt tranporter<br />
dtd b3887 1.2548 0.0380 D-Tyr-tRNATyr deacylase<br />
elaC b2268 0.6406 0.0465 binuclear zinc phosphodiesterase monomer<br />
emrE b0543 0.4603 0.0035 EmrE SMR transporter<br />
emrK b2368 0.5558 0.0366 EmrK putative membrane fusion protein<br />
emrY b2367 0.1336 0.0000 EmrY putative multidrug MFS transporter<br />
envC b3613 1.5602 0.0371 EnvC murein hydrolase<br />
essQ b1556 4.4414 0.0125 hypothetical protein<br />
eutB b2441 0.7106 0.0143 ethanolamine ammonia-lyase, probable regulatory subunit<br />
eutD b2458 0.4881 0.0053 putative phosphate acetyltransferase, ethanolamine utilization<br />
eutN b2456 0.5116 0.0035 putative detox protein, ethanolamine utilization<br />
GDP-4-dehydro-6-deoxy-D-mannose epimerase / GDP-4-dehydro-6-Ldeoxygalactose<br />
fcl b2052 0.0327 0.0006<br />
reductase<br />
outer membrane receptor; citrate-dependent iron transport, outer membrane<br />
fecA b4291 0.6549 0.0354 receptor<br />
fecB b4290 0.6183 0.0038 ferric dicitrate uptake system<br />
fhlA b2731 1.2443 0.0006 FhlA-Formate transcriptional activator<br />
fhuC b0151 2.0062 0.0140 ferrichrome uptake system<br />
stationary-phase protein with possible role in p-aminobenzoate or folate<br />
fic b3361 1.3138 0.0372 biosynthesis<br />
fimA b4314 0.0526 0.0007 major type 1 subunit fimbrin (pilin)<br />
fimB b4312 0.0776 0.0001 regulator for fimA<br />
fimD b4317 0.3019 0.0243 outer membrane protein; export and assembly <strong>of</strong> type 1 fimbriae, interrupted<br />
fimE b4313 0.0565 0.0001 regulator for fimA<br />
fimG b4319 0.5755 0.0041 fimbrial morphology<br />
fimI b4315 0.0712 0.0005 fimbrial protein<br />
fixB b0042 0.3451 0.0185 probable flavoprotein subunit required for anaerobic carnitine metabolism<br />
flavoprotein (electron transport), possibly involved in anaerobic carnitine<br />
fixC b0043 0.4760 0.0273 metabolism<br />
flgA b1072 27.1534 0.0010 flagellar biosynthesis; assembly <strong>of</strong> basal-body periplasmic P ring<br />
flgB b1073 24.5528 0.0038 flagellar basal-body rod protein FlgB<br />
flgC b1074 14.9393 0.0009 flagellar basal-body rod protein FlgC<br />
flgD b1075 16.5742 0.0019 flagellar biosynthesis, initiation <strong>of</strong> hook assembly<br />
flgE b1076 9.2717 0.0007 flagellar hook protein FlgE<br />
flgF b1077 6.1174 0.0007 flagellar basal-body rod protein FlgF<br />
flgG b1078 21.9901 0.0010 flagellar basal-body rod protein FlgG<br />
flagellar L-ring protein FlgH; basal-body outer-membrane L (lipopolysaccharide<br />
flgH b1079 9.7694 0.0040 layer) ring protein<br />
flgI b1080 15.6181 0.0055 flagellar P-ring protein FlgI<br />
flgJ b1081 6.3428 0.0005 FlgJ<br />
flgK b1082 4.5952 0.0289 flagellar biosynthesis, hook-filament junction protein 1<br />
flgM b1071 28.5501 0.0060 anti-FliA (anti-sigma) factor; also known as RflB protein<br />
flgN b1070 7.0812 0.0008 flagellar biosynthesis protein FlgN<br />
flhA b1879 2.5230 0.0463 flagellar biosynthesis protein FlhA<br />
flhB b1880 10.7222 0.0025 flagellar biosynthesis protein FlhB<br />
169