Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
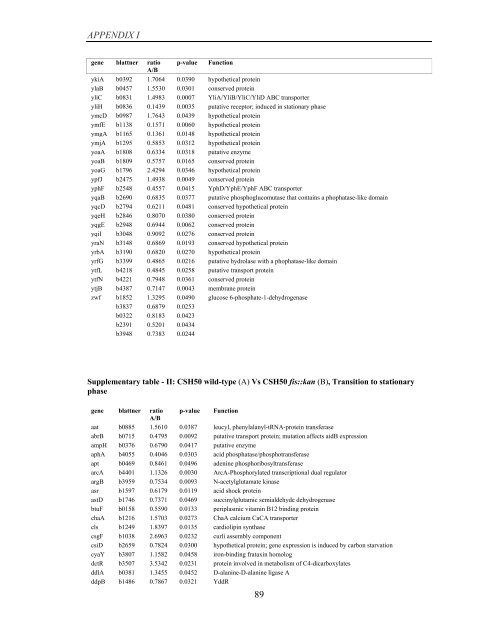
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
ykiA b0392 1.7064 0.0390 hypothetical protein<br />
ylaB b0457 1.5530 0.0301 conserved protein<br />
yliC b0831 1.4983 0.0007 YliA/YliB/YliC/YliD ABC transporter<br />
yliH b0836 0.1439 0.0035 putative receptor; induced in stationary phase<br />
ymcD b0987 1.7643 0.0439 hypothetical protein<br />
ymfE b1138 0.1571 0.0060 hypothetical protein<br />
ymgA b1165 0.1361 0.0148 hypothetical protein<br />
ymjA b1295 0.5853 0.0312 hypothetical protein<br />
yoaA b1808 0.6334 0.0318 putative enzyme<br />
yoaB b1809 0.5757 0.0165 conserved protein<br />
yoaG b1796 2.4294 0.0346 hypothetical protein<br />
ypfJ b2475 1.4938 0.0049 conserved protein<br />
yphF b2548 0.4557 0.0415 YphD/YphE/YphF ABC transporter<br />
yqaB b2690 0.6835 0.0377 putative phosphoglucomutase that contains a phophatase-like domain<br />
yqcD b2794 0.6211 0.0481 conserved hypothetical protein<br />
yqeH b2846 0.8070 0.0380 conserved protein<br />
yqgE b2948 0.6944 0.0062 conserved protein<br />
yqiI b3048 0.9092 0.0276 conserved protein<br />
yraN b3148 0.6869 0.0193 conserved hypothetical protein<br />
yrbA b3190 0.6820 0.0270 hypothetical protein<br />
yrfG b3399 0.4865 0.0216 putative hydrolase with a phophatase-like domain<br />
ytfL b4218 0.4845 0.0258 putative transport protein<br />
ytfN b4221 0.7948 0.0361 conserved protein<br />
ytjB b4387 0.7147 0.0043 membrane protein<br />
zwf b1852 1.3295 0.0490 glucose 6-phosphate-1-dehydrogenase<br />
b3837 0.6879 0.0253<br />
b0322 0.8183 0.0423<br />
b2391 0.5201 0.0434<br />
b3948 0.7383 0.0244<br />
Supplementary table - II: CSH50 wild-type (A) Vs CSH50 fis::kan (B), Transition to stationary<br />
phase<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
aat b0885 1.5610 0.0387 leucyl, phenylalanyl-tRNA-protein transferase<br />
abrB b0715 0.4795 0.0092 putative transport protein; mutation affects aidB <strong>expression</strong><br />
ampH b0376 0.6790 0.0417 putative enzyme<br />
aphA b4055 0.4046 0.0303 acid phosphatase/phosphotransferase<br />
apt b0469 0.8461 0.0496 adenine phosphoribosyltransferase<br />
arcA b4401 1.1326 0.0030 ArcA-Phosphorylated transcriptional dual regulator<br />
argB b3959 0.7534 0.0093 N-acetylglutamate kinase<br />
asr b1597 0.6179 0.0119 acid shock protein<br />
astD b1746 0.7371 0.0469 succinylglutamic semialdehyde dehydrogenase<br />
btuF b0158 0.5590 0.0133 periplasmic vitamin B12 binding protein<br />
chaA b1216 1.5703 0.0273 ChaA calcium CaCA transporter<br />
cls b1249 1.8397 0.0135 cardiolipin synthase<br />
csgF b1038 2.6963 0.0232 curli assembly component<br />
csiD b2659 0.7824 0.0300 hypothetical protein; <strong>gene</strong> <strong>expression</strong> is induced <strong>by</strong> carbon starvation<br />
cyaY b3807 1.1582 0.0458 iron-binding frataxin homolog<br />
dctR b3507 3.5342 0.0231 protein involved in metabolism <strong>of</strong> C4-dicarboxylates<br />
ddlA b0381 1.3455 0.0452 D-alanine-D-alanine ligase A<br />
ddpB b1486 0.7867 0.0321 YddR<br />
89