Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
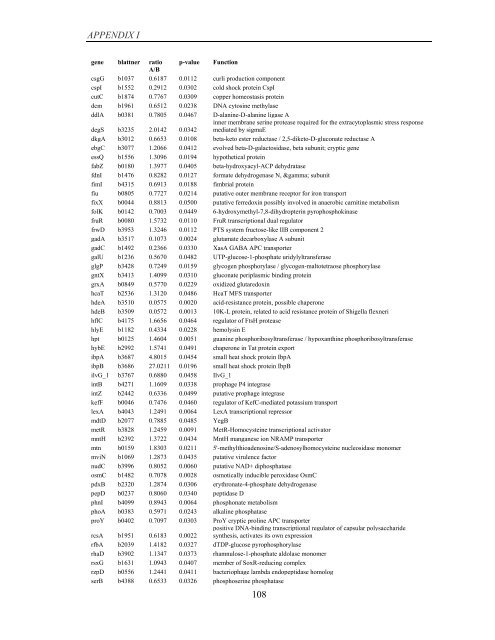
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
csgG b1037 0.6187 0.0112 curli production component<br />
cspI b1552 0.2912 0.0302 cold shock protein CspI<br />
cutC b1874 0.7767 0.0309 copper homeostasis protein<br />
dcm b1961 0.6512 0.0238 DNA cytosine methylase<br />
ddlA b0381 0.7805 0.0467 D-alanine-D-alanine ligase A<br />
degS b3235 2.0142 0.0342<br />
inner membrane serine protease required for the extracytoplasmic stress response<br />
mediated <strong>by</strong> sigmaE<br />
dkgA b3012 0.6653 0.0108 beta-keto ester reductase / 2,5-diketo-D-gluconate reductase A<br />
ebgC b3077 1.2066 0.0412 evolved beta-D-galactosidase, beta subunit; cryptic <strong>gene</strong><br />
essQ b1556 1.3096 0.0194 hypothetical protein<br />
fabZ b0180 1.3977 0.0405 beta-hydroxyacyl-ACP dehydratase<br />
fdnI b1476 0.8282 0.0127 formate dehydrogenase N, γ subunit<br />
fimI b4315 0.6913 0.0188 fimbrial protein<br />
fiu b0805 0.7727 0.0214 putative outer membrane receptor for iron transport<br />
fixX b0044 0.8813 0.0500 putative ferredoxin possibly involved in anaerobic carnitine metabolism<br />
folK b0142 0.7003 0.0449 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase<br />
fruR b0080 1.5732 0.0110 FruR transcriptional dual regulator<br />
frwD b3953 1.3246 0.0112 PTS system fructose-like IIB component 2<br />
gadA b3517 0.1073 0.0024 glutamate decarboxylase A subunit<br />
gadC b1492 0.2366 0.0330 XasA GABA APC transporter<br />
galU b1236 0.5670 0.0482 UTP-glucose-1-phosphate uridylyltransferase<br />
glgP b3428 0.7249 0.0159 glycogen phosphorylase / glycogen-maltotetraose phosphorylase<br />
gntX b3413 1.4099 0.0310 gluconate periplasmic binding protein<br />
grxA b0849 0.5770 0.0229 oxidized glutaredoxin<br />
hcaT b2536 1.3120 0.0486 HcaT MFS transporter<br />
hdeA b3510 0.0575 0.0020 acid-resistance protein, possible chaperone<br />
hdeB b3509 0.0572 0.0013 10K-L protein, related to acid resistance protein <strong>of</strong> Shigella flexneri<br />
hflC b4175 1.6656 0.0464 regulator <strong>of</strong> FtsH protease<br />
hlyE b1182 0.4334 0.0228 hemolysin E<br />
hpt b0125 1.4604 0.0051 guanine phosphoribosyltransferase / hypoxanthine phosphoribosyltransferase<br />
hybE b2992 1.5741 0.0491 chaperone in Tat protein export<br />
ibpA b3687 4.8015 0.0454 small heat shock protein IbpA<br />
ibpB b3686 27.0211 0.0196 small heat shock protein IbpB<br />
ilvG_1 b3767 0.6880 0.0458 IlvG_1<br />
intB b4271 1.1609 0.0338 prophage P4 integrase<br />
intZ b2442 0.6336 0.0499 putative prophage integrase<br />
kefF b0046 0.7476 0.0460 regulator <strong>of</strong> KefC-mediated potassium transport<br />
lexA b4043 1.2491 0.0064 LexA transcriptional repressor<br />
mdtD b2077 0.7885 0.0485 YegB<br />
metR b3828 1.2459 0.0091 MetR-Homocysteine transcriptional activator<br />
mntH b2392 1.3722 0.0434 MntH manganese ion NRAMP transporter<br />
mtn b0159 1.8303 0.0211 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase monomer<br />
mviN b1069 1.2873 0.0435 putative virulence factor<br />
nudC b3996 0.8052 0.0060 putative NAD+ diphosphatase<br />
osmC b1482 0.7078 0.0028 osmotically inducible peroxidase OsmC<br />
pdxB b2320 1.2874 0.0306 erythronate-4-phosphate dehydrogenase<br />
pepD b0237 0.8060 0.0340 peptidase D<br />
phnI b4099 0.8943 0.0064 phosphonate metabolism<br />
phoA b0383 0.5971 0.0243 alkaline phosphatase<br />
proY b0402 0.7097 0.0303 ProY cryptic proline APC transporter<br />
rcsA b1951 0.6183 0.0022<br />
positive DNA-binding transcriptional regulator <strong>of</strong> capsular polysaccharide<br />
synthesis, activates its own <strong>expression</strong><br />
rfbA b2039 1.4182 0.0327 dTDP-glucose pyrophosphorylase<br />
rhaD b3902 1.1347 0.0373 rhamnulose-1-phosphate aldolase monomer<br />
rsxG b1631 1.0943 0.0407 member <strong>of</strong> SoxR-reducing complex<br />
rzpD b0556 1.2441 0.0411 bacteriophage lambda endopeptidase homolog<br />
serB b4388 0.6533 0.0326 phosphoserine phosphatase<br />
108