Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
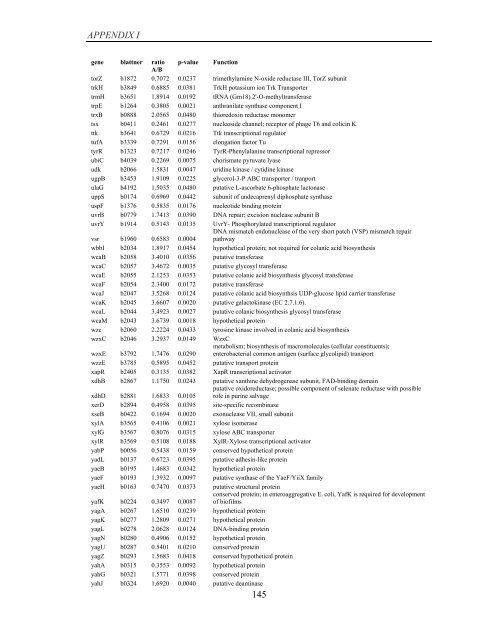
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
torZ b1872 0.7072 0.0237 trimethylamine N-oxide reductase III, TorZ subunit<br />
trkH b3849 0.6885 0.0381 TrkH potassium ion Trk Transporter<br />
trmH b3651 1.8914 0.0192 tRNA (Gm18) 2'-O-methyltransferase<br />
trpE b1264 0.3805 0.0021 anthranilate synthase component I<br />
trxB b0888 2.0565 0.0480 thioredoxin reductase monomer<br />
tsx b0411 0.2461 0.0277 nucleoside channel; receptor <strong>of</strong> phage T6 and colicin K<br />
ttk b3641 0.6729 0.0216 Ttk transcriptional regulator<br />
tufA b3339 0.7291 0.0156 elongation factor Tu<br />
tyrR b1323 0.7217 0.0246 TyrR-Phenylalanine transcriptional repressor<br />
ubiC b4039 0.2269 0.0075 chorismate pyruvate lyase<br />
udk b2066 1.5831 0.0047 uridine kinase / cytidine kinase<br />
ugpB b3453 1.9109 0.0225 glycerol-3-P ABC transporter / tranport<br />
ulaG b4192 1.5035 0.0480 putative L-ascorbate 6-phosphate lactonase<br />
uppS b0174 0.6969 0.0442 subunit <strong>of</strong> undecaprenyl diphosphate synthase<br />
uspF b1376 0.5835 0.0176 nucleotide binding protein<br />
uvrB b0779 1.7413 0.0390 DNA repair; excision nuclease subunit B<br />
uvrY b1914 0.5143 0.0135 UvrY- Phosphorylated transcriptional regulator<br />
vsr b1960 0.6583 0.0004<br />
DNA mismatch endonuclease <strong>of</strong> the very short patch (VSP) mismatch repair<br />
pathway<br />
wbbI b2034 1.8917 0.0454 hypothetical protein; not required for colanic acid biosynthesis<br />
wcaB b2058 3.4010 0.0356 putative transferase<br />
wcaC b2057 3.4672 0.0035 putative glycosyl transferase<br />
wcaE b2055 2.1253 0.0353 putative colanic acid biosynthesis glycosyl transferase<br />
wcaF b2054 2.3400 0.0172 putative transferase<br />
wcaJ b2047 3.5268 0.0124 putative colanic acid biosynthsis UDP-glucose lipid carrier transferase<br />
wcaK b2045 3.6607 0.0020 putative galactokinase (EC 2.7.1.6).<br />
wcaL b2044 3.4923 0.0027 putative colanic biosynthesis glycosyl transferase<br />
wcaM b2043 3.6739 0.0018 hypothetical protein<br />
wzc b2060 2.2224 0.0433 tyrosine kinase involved in colanic acid biosynthesis<br />
wzxC b2046 3.2937 0.0149 WzxC<br />
wzxE b3792 1.7476 0.0290<br />
metabolism; biosynthesis <strong>of</strong> macromolecules (cellular constituents);<br />
enterobacterial common antigen (surface glycolipid) transport<br />
wzzE b3785 0.5895 0.0452 putative transport protein<br />
xapR b2405 0.3135 0.0382 XapR transcriptional activator<br />
xdhB b2867 1.1750 0.0243 putative xanthine dehydrogenase subunit, FAD-binding domain<br />
xdhD b2881 1.6833 0.0105<br />
putative oxidoreductase; possible component <strong>of</strong> selenate reductase with possible<br />
role in purine salvage<br />
xerD b2894 0.4958 0.0395 site-specific recombinase<br />
xseB b0422 0.1694 0.0020 exonuclease VII, small subunit<br />
xylA b3565 0.4106 0.0021 xylose isomerase<br />
xylG b3567 0.8076 0.0315 xylose ABC transporter<br />
xylR b3569 0.5108 0.0188 XylR-Xylose transcriptional activator<br />
yabP b0056 0.5438 0.0159 conserved hypothetical protein<br />
yadL b0137 0.6723 0.0395 putative adhesin-like protein<br />
yaeB b0195 1.4683 0.0342 hypothetical protein<br />
yaeF b0193 1.3932 0.0097 putative synthase <strong>of</strong> the YaeF/YiiX family<br />
yaeH b0163 0.7470 0.0373 putative structural protein<br />
yafK b0224 0.3497 0.0087<br />
conserved protein; in enteroaggregative E. coli, YafK is required for development<br />
<strong>of</strong> bi<strong>of</strong>ilms<br />
yagA b0267 1.6510 0.0239 hypothetical protein<br />
yagK b0277 1.2809 0.0271 hypothetical protein<br />
yagL b0278 2.0628 0.0124 DNA-binding protein<br />
yagN b0280 0.4906 0.0152 hypothetical protein<br />
yagU b0287 0.5401 0.0210 conserved protein<br />
yagZ b0293 1.5683 0.0418 conserved hypothetical protein<br />
yahA b0315 0.3553 0.0092 hypothetical protein<br />
yahG b0321 1.5771 0.0398 conserved protein<br />
yahJ b0324 1.6920 0.0040 putative deaminase<br />
145