Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
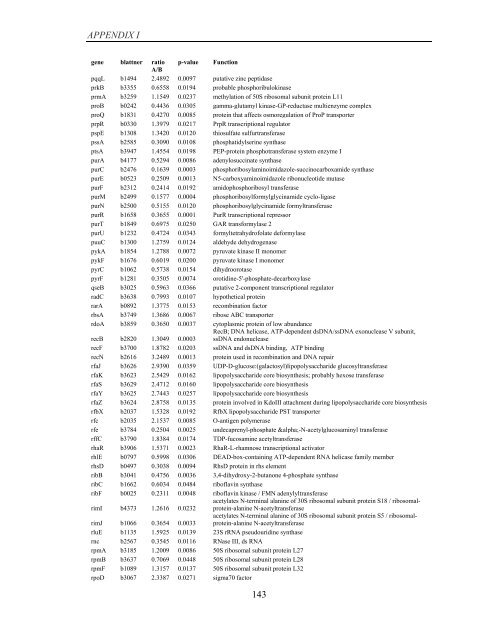
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
pqqL b1494 2.4892 0.0097 putative zinc peptidase<br />
prkB b3355 0.6558 0.0194 probable phosphoribulokinase<br />
prmA b3259 1.1549 0.0237 methylation <strong>of</strong> 50S ribosomal subunit protein L11<br />
proB b0242 0.4436 0.0305 gamma-glutamyl kinase-GP-reductase multienzyme complex<br />
proQ b1831 0.4270 0.0085 protein that affects osmo<strong>regulation</strong> <strong>of</strong> ProP transporter<br />
prpR b0330 1.3979 0.0217 PrpR transcriptional regulator<br />
pspE b1308 1.3420 0.0120 thiosulfate sulfurtransferase<br />
pssA b2585 0.3090 0.0108 phosphatidylserine synthase<br />
ptsA b3947 1.4554 0.0198 PEP-protein phosphotransferase system enzyme I<br />
purA b4177 0.5294 0.0086 adenylosuccinate synthase<br />
purC b2476 0.1639 0.0003 phosphoribosylaminoimidazole-succinocarboxamide synthase<br />
purE b0523 0.2509 0.0013 N5-carboxyaminoimidazole ribonucleotide mutase<br />
purF b2312 0.2414 0.0192 amidophosphoribosyl transferase<br />
purM b2499 0.1577 0.0004 phosphoribosylformylglycinamide cyclo-ligase<br />
purN b2500 0.5155 0.0120 phosphoribosylglycinamide formyltransferase<br />
purR b1658 0.3655 0.0001 PurR transcriptional repressor<br />
purT b1849 0.6975 0.0250 GAR transformylase 2<br />
purU b1232 0.4724 0.0343 formyltetrahydr<strong>of</strong>olate deformylase<br />
puuC b1300 1.2759 0.0124 aldehyde dehydrogenase<br />
pykA b1854 1.2788 0.0072 pyruvate kinase II monomer<br />
pykF b1676 0.6019 0.0200 pyruvate kinase I monomer<br />
pyrC b1062 0.5738 0.0154 dihydroorotase<br />
pyrF b1281 0.3505 0.0074 orotidine-5'-phosphate-decarboxylase<br />
qseB b3025 0.5963 0.0366 putative 2-component transcriptional regulator<br />
radC b3638 0.7993 0.0107 hypothetical protein<br />
rarA b0892 1.3775 0.0153 recombination factor<br />
rbsA b3749 1.3686 0.0067 ribose ABC transporter<br />
rdoA b3859 0.3650 0.0037 cytoplasmic protein <strong>of</strong> low abundance<br />
recB b2820 1.3049 0.0003<br />
RecB; DNA helicase, ATP-dependent dsDNA/ssDNA exonuclease V subunit,<br />
ssDNA endonuclease<br />
recF b3700 1.8782 0.0203 ssDNA and dsDNA binding, ATP binding<br />
recN b2616 3.2489 0.0013 protein used in recombination and DNA repair<br />
rfaJ b3626 2.9390 0.0359 UDP-D-glucose:(galactosyl)lipopolysaccharide glucosyltransferase<br />
rfaK b3623 2.5429 0.0162 lipopolysaccharide core biosynthesis; probably hexose transferase<br />
rfaS b3629 2.4712 0.0160 lipopolysaccharide core biosynthesis<br />
rfaY b3625 2.7443 0.0257 lipopolysaccharide core biosynthesis<br />
rfaZ b3624 2.8758 0.0135 protein involved in KdoIII attachment during lipopolysaccharide core biosynthesis<br />
rfbX b2037 1.5328 0.0192 RfbX lipopolysaccharide PST transporter<br />
rfc b2035 2.1537 0.0085 O-antigen polymerase<br />
rfe b3784 0.2504 0.0025 undecaprenyl-phosphate α-N-acetylglucosaminyl transferase<br />
rffC b3790 1.8384 0.0174 TDP-fucosamine acetyltransferase<br />
rhaR b3906 1.5371 0.0023 RhaR-L-rhamnose transcriptional activator<br />
rhlE b0797 0.5998 0.0306 DEAD-box-containing ATP-dependent RNA helicase family member<br />
rhsD b0497 0.3038 0.0094 RhsD protein in rhs element<br />
ribB b3041 0.4756 0.0036 3,4-dihydroxy-2-butanone 4-phosphate synthase<br />
ribC b1662 0.6034 0.0484 rib<strong>of</strong>lavin synthase<br />
ribF b0025 0.2311 0.0048 rib<strong>of</strong>lavin kinase / FMN adenylyltransferase<br />
rimI b4373 1.2616 0.0232<br />
acetylates N-terminal alanine <strong>of</strong> 30S ribosomal subunit protein S18 / ribosomalprotein-alanine<br />
N-acetyltransferase<br />
rimJ b1066 0.3654 0.0033<br />
acetylates N-terminal alanine <strong>of</strong> 30S ribosomal subunit protein S5 / ribosomalprotein-alanine<br />
N-acetyltransferase<br />
rluE b1135 1.5925 0.0139 23S rRNA pseudouridine synthase<br />
rnc b2567 0.3545 0.0116 RNase III, ds RNA<br />
rpmA b3185 1.2009 0.0086 50S ribosomal subunit protein L27<br />
rpmB b3637 0.7069 0.0448 50S ribosomal subunit protein L28<br />
rpmF b1089 1.3157 0.0137 50S ribosomal subunit protein L32<br />
rpoD b3067 2.3387 0.0271 sigma70 factor<br />
143