Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
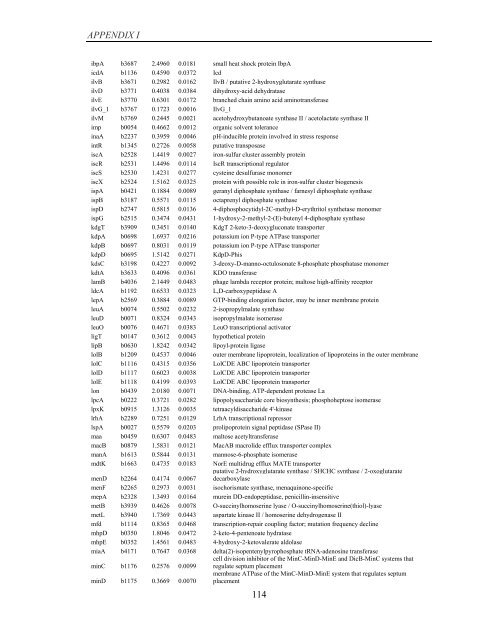
APPENDIX I<br />
ibpA b3687 2.4960 0.0181 small heat shock protein IbpA<br />
icdA b1136 0.4590 0.0372 Icd<br />
ilvB b3671 0.2982 0.0162 IlvB / putative 2-hydroxyglutarate synthase<br />
ilvD b3771 0.4038 0.0384 dihydroxy-acid dehydratase<br />
ilvE b3770 0.6301 0.0172 branched chain amino acid aminotransferase<br />
ilvG_1 b3767 0.1723 0.0016 IlvG_1<br />
ilvM b3769 0.2445 0.0021 acetohydroxybutanoate synthase II / acetolactate synthase II<br />
imp b0054 0.4662 0.0012 organic solvent tolerance<br />
inaA b2237 0.3959 0.0046 pH-inducible protein involved in stress response<br />
intR b1345 0.2726 0.0058 putative transposase<br />
iscA b2528 1.4419 0.0027 iron-sulfur cluster assembly protein<br />
iscR b2531 1.4496 0.0114 IscR transcriptional regulator<br />
iscS b2530 1.4231 0.0277 cysteine desulfurase monomer<br />
iscX b2524 1.5162 0.0325 protein with possible role in iron-sulfur cluster bio<strong>gene</strong>sis<br />
ispA b0421 0.1884 0.0089 geranyl diphosphate synthase / farnesyl diphosphate synthase<br />
ispB b3187 0.5571 0.0115 octaprenyl diphosphate synthase<br />
ispD b2747 0.5815 0.0136 4-diphosphocytidyl-2C-methyl-D-erythritol synthetase monomer<br />
ispG b2515 0.3474 0.0431 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase<br />
kdgT b3909 0.3451 0.0140 KdgT 2-keto-3-deoxygluconate transporter<br />
kdpA b0698 1.6937 0.0216 potassium ion P-type ATPase transporter<br />
kdpB b0697 0.8031 0.0119 potassium ion P-type ATPase transporter<br />
kdpD b0695 1.5142 0.0271 KdpD-Phis<br />
kdsC b3198 0.4227 0.0092 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase monomer<br />
kdtA b3633 0.4096 0.0361 KDO transferase<br />
lamB b4036 2.1449 0.0483 phage lambda receptor protein; maltose high-affinity receptor<br />
ldcA b1192 0.6533 0.0323 L,D-carboxypeptidase A<br />
lepA b2569 0.3884 0.0089 GTP-binding elongation factor, may be inner membrane protein<br />
leuA b0074 0.5502 0.0232 2-isopropylmalate synthase<br />
leuD b0071 0.8324 0.0343 isopropylmalate isomerase<br />
leuO b0076 0.4671 0.0383 LeuO transcriptional activator<br />
ligT b0147 0.3612 0.0043 hypothetical protein<br />
lipB b0630 1.8242 0.0342 lipoyl-protein ligase<br />
lolB b1209 0.4537 0.0046 outer membrane lipoprotein, localization <strong>of</strong> lipoproteins in the outer membrane<br />
lolC b1116 0.4315 0.0356 LolCDE ABC lipoprotein transporter<br />
lolD b1117 0.6023 0.0038 LolCDE ABC lipoprotein transporter<br />
lolE b1118 0.4199 0.0393 LolCDE ABC lipoprotein transporter<br />
lon b0439 2.0180 0.0071 DNA-binding, ATP-dependent protease La<br />
lpcA b0222 0.3721 0.0282 lipopolysaccharide core biosynthesis; phosphoheptose isomerase<br />
lpxK b0915 1.3126 0.0035 tetraacyldisaccharide 4'-kinase<br />
lrhA b2289 0.7251 0.0129 LrhA transcriptional repressor<br />
lspA b0027 0.5579 0.0203 prolipoprotein signal peptidase (SPase II)<br />
maa b0459 0.6307 0.0483 maltose acetyltransferase<br />
macB b0879 1.5831 0.0121 MacAB macrolide efflux transporter complex<br />
manA b1613 0.5844 0.0131 mannose-6-phosphate isomerase<br />
mdtK b1663 0.4735 0.0183 NorE multidrug efflux MATE transporter<br />
menD b2264 0.4174 0.0067<br />
putative 2-hydroxyglutarate synthase / SHCHC synthase / 2-oxoglutarate<br />
decarboxylase<br />
menF b2265 0.2973 0.0031 isochorismate synthase, menaquinone-specific<br />
mepA b2328 1.3493 0.0164 murein DD-endopeptidase, penicillin-insensitive<br />
metB b3939 0.4626 0.0078 O-succinylhomoserine lyase / O-succinylhomoserine(thiol)-lyase<br />
metL b3940 1.7369 0.0443 aspartate kinase II / homoserine dehydrogenase II<br />
mfd b1114 0.8365 0.0468 transcription-repair coupling factor; mutation frequency decline<br />
mhpD b0350 1.8046 0.0472 2-keto-4-pentenoate hydratase<br />
mhpE b0352 1.4561 0.0483 4-hydroxy-2-ketovalerate aldolase<br />
miaA b4171 0.7647 0.0368 delta(2)-isopentenylpyrophosphate tRNA-adenosine transferase<br />
minC b1176 0.2576 0.0099<br />
cell division inhibitor <strong>of</strong> the MinC-MinD-MinE and DicB-MinC systems that<br />
regulate septum placement<br />
membrane ATPase <strong>of</strong> the MinC-MinD-MinE system that regulates septum<br />
minD b1175 0.3669 0.0070 placement<br />
114