Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
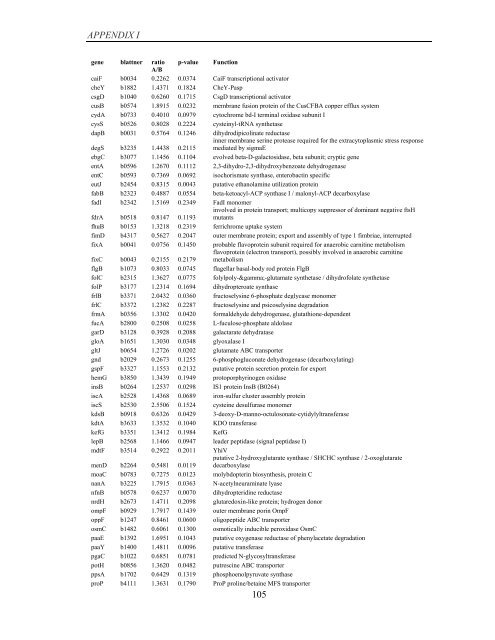
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
caiF b0034 0.2262 0.0374 CaiF transcriptional activator<br />
cheY b1882 1.4371 0.1824 CheY-Pasp<br />
csgD b1040 0.6260 0.1715 CsgD transcriptional activator<br />
cusB b0574 1.8915 0.0232 membrane fusion protein <strong>of</strong> the CusCFBA copper efflux system<br />
cydA b0733 0.4010 0.0979 cytochrome bd-I terminal oxidase subunit I<br />
cysS b0526 0.8028 0.2224 cysteinyl-tRNA synthetase<br />
dapB b0031 0.5764 0.1246 dihydrodipicolinate reductase<br />
degS b3235 1.4438 0.2115<br />
inner membrane serine protease required for the extracytoplasmic stress response<br />
mediated <strong>by</strong> sigmaE<br />
ebgC b3077 1.1456 0.1104 evolved beta-D-galactosidase, beta subunit; cryptic <strong>gene</strong><br />
entA b0596 1.2670 0.1112 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase<br />
entC b0593 0.7369 0.0692 isochorismate synthase, enterobactin specific<br />
eutJ b2454 0.8315 0.0043 putative ethanolamine utilization protein<br />
fabB b2323 0.4887 0.0554 beta-ketoacyl-ACP synthase I / malonyl-ACP decarboxylase<br />
fadI b2342 1.5169 0.2349 FadI monomer<br />
fdrA b0518 0.8147 0.1193<br />
involved in protein transport; multicopy suppressor <strong>of</strong> dominant negative ftsH<br />
mutants<br />
fhuB b0153 1.3218 0.2319 ferrichrome uptake system<br />
fimD b4317 0.5627 0.2047 outer membrane protein; export and assembly <strong>of</strong> type 1 fimbriae, interrupted<br />
fixA b0041 0.0756 0.1450 probable flavoprotein subunit required for anaerobic carnitine metabolism<br />
fixC b0043 0.2155 0.2179<br />
flavoprotein (electron transport), possibly involved in anaerobic carnitine<br />
metabolism<br />
flgB b1073 0.8033 0.0745 flagellar basal-body rod protein FlgB<br />
folC b2315 1.3627 0.0775 folylpoly-γ-glutamate synthetase / dihydr<strong>of</strong>olate synthetase<br />
folP b3177 1.2314 0.1694 dihydropteroate synthase<br />
frlB b3371 2.0432 0.0360 fructoselysine 6-phosphate deglycase monomer<br />
frlC b3372 1.2382 0.2287 fructoselysine and psicoselysine degradation<br />
frmA b0356 1.3302 0.0420 formaldehyde dehydrogenase, glutathione-dependent<br />
fucA b2800 0.2508 0.0258 L-fuculose-phosphate aldolase<br />
garD b3128 0.3928 0.2088 galactarate dehydratase<br />
gloA b1651 1.3030 0.0348 glyoxalase I<br />
gltJ b0654 1.2726 0.0202 glutamate ABC transporter<br />
gnd b2029 0.2673 0.1255 6-phosphogluconate dehydrogenase (decarboxylating)<br />
gspF b3327 1.1553 0.2132 putative protein secretion protein for export<br />
hemG b3850 1.3439 0.1949 protoporphyrinogen oxidase<br />
insB b0264 1.2537 0.0298 IS1 protein InsB (B0264)<br />
iscA b2528 1.4368 0.0689 iron-sulfur cluster assembly protein<br />
iscS b2530 2.5506 0.1524 cysteine desulfurase monomer<br />
kdsB b0918 0.6326 0.0429 3-deoxy-D-manno-octulosonate-cytidylyltransferase<br />
kdtA b3633 1.3532 0.1040 KDO transferase<br />
kefG b3351 1.3412 0.1984 KefG<br />
lepB b2568 1.1466 0.0947 leader peptidase (signal peptidase I)<br />
mdtF b3514 0.2922 0.2011 YhiV<br />
menD b2264 0.5481 0.0119<br />
putative 2-hydroxyglutarate synthase / SHCHC synthase / 2-oxoglutarate<br />
decarboxylase<br />
moaC b0783 0.7275 0.0123 molybdopterin biosynthesis, protein C<br />
nanA b3225 1.7915 0.0363 N-acetylneuraminate lyase<br />
nfnB b0578 0.6237 0.0070 dihydropteridine reductase<br />
nrdH b2673 1.4711 0.2098 glutaredoxin-like protein; hydrogen donor<br />
ompF b0929 1.7917 0.1439 outer membrane porin OmpF<br />
oppF b1247 0.8461 0.0600 oligopeptide ABC transporter<br />
osmC b1482 0.6061 0.1300 osmotically inducible peroxidase OsmC<br />
paaE b1392 1.6951 0.1043 putative oxygenase reductase <strong>of</strong> phenylacetate degradation<br />
paaY b1400 1.4811 0.0096 putative transferase<br />
pgaC b1022 0.6851 0.0781 predicted N-glycosyltransferase<br />
potH b0856 1.3620 0.0482 putrescine ABC transporter<br />
ppsA b1702 0.6429 0.1319 phosphoenolpyruvate synthase<br />
proP b4111 1.3631 0.1790 ProP proline/betaine MFS transporter<br />
105