Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
Coordinated regulation of gene expression by E ... - Jacobs University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
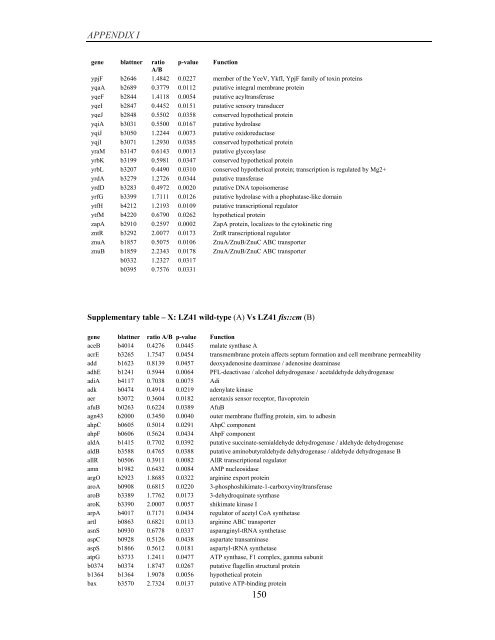
APPENDIX I<br />
<strong>gene</strong> blattner ratio p-value Function<br />
A/B<br />
ypjF b2646 1.4842 0.0227 member <strong>of</strong> the YeeV, YkfI, YpjF family <strong>of</strong> toxin proteins<br />
yqaA b2689 0.3779 0.0112 putative integral membrane protein<br />
yqeF b2844 1.4118 0.0054 putative acyltransferase<br />
yqeI b2847 0.4452 0.0151 putative sensory transducer<br />
yqeJ b2848 0.5502 0.0358 conserved hypothetical protein<br />
yqiA b3031 0.5500 0.0167 putative hydrolase<br />
yqiJ b3050 1.2244 0.0073 putative oxidoreductase<br />
yqjI b3071 1.2930 0.0385 conserved hypothetical protein<br />
yraM b3147 0.6143 0.0013 putative glycosylase<br />
yrbK b3199 0.5981 0.0347 conserved hypothetical protein<br />
yrbL b3207 0.4490 0.0310 conserved hypothetical protein; transcription is regulated <strong>by</strong> Mg2+<br />
yrdA b3279 1.2726 0.0344 putative transferase<br />
yrdD b3283 0.4972 0.0020 putative DNA topoisomerase<br />
yrfG b3399 1.7111 0.0126 putative hydrolase with a phophatase-like domain<br />
ytfH b4212 1.2193 0.0109 putative transcriptional regulator<br />
ytfM b4220 0.6790 0.0262 hypothetical protein<br />
zapA b2910 0.2597 0.0002 ZapA protein, localizes to the cytokinetic ring<br />
zntR b3292 2.0077 0.0173 ZntR transcriptional regulator<br />
znuA b1857 0.5075 0.0106 ZnuA/ZnuB/ZnuC ABC transporter<br />
znuB b1859 2.2343 0.0178 ZnuA/ZnuB/ZnuC ABC transporter<br />
b0332 1.2327 0.0317<br />
b0395 0.7576 0.0331<br />
Supplementary table – X: LZ41 wild-type (A) Vs LZ41 fis::cm (B)<br />
<strong>gene</strong> blattner ratio A/B p-value Function<br />
aceB b4014 0.4276 0.0445 malate synthase A<br />
acrE b3265 1.7547 0.0454 transmembrane protein affects septum formation and cell membrane permeability<br />
add b1623 0.8139 0.0457 deoxyadenosine deaminase / adenosine deaminase<br />
adhE b1241 0.5944 0.0064 PFL-deactivase / alcohol dehydrogenase / acetaldehyde dehydrogenase<br />
adiA b4117 0.7038 0.0075 Adi<br />
adk b0474 0.4914 0.0219 adenylate kinase<br />
aer b3072 0.3604 0.0182 aerotaxis sensor receptor, flavoprotein<br />
afuB b0263 0.6224 0.0389 AfuB<br />
agn43 b2000 0.3450 0.0040 outer membrane fluffing protein, sim. to adhesin<br />
ahpC b0605 0.5014 0.0291 AhpC component<br />
ahpF b0606 0.5624 0.0434 AhpF component<br />
aldA b1415 0.7702 0.0392 putative succinate-semialdehyde dehydrogenase / aldehyde dehydrogenase<br />
aldB b3588 0.4765 0.0388 putative aminobutyraldehyde dehydrogenase / aldehyde dehydrogenase B<br />
allR b0506 0.3911 0.0082 AllR transcriptional regulator<br />
amn b1982 0.6432 0.0084 AMP nucleosidase<br />
argO b2923 1.8685 0.0322 arginine export protein<br />
aroA b0908 0.6815 0.0220 3-phosphoshikimate-1-carboxyvinyltransferase<br />
aroB b3389 1.7762 0.0173 3-dehydroquinate synthase<br />
aroK b3390 2.0007 0.0057 shikimate kinase I<br />
arpA b4017 0.7171 0.0434 regulator <strong>of</strong> acetyl CoA synthetase<br />
artI b0863 0.6821 0.0113 arginine ABC transporter<br />
asnS b0930 0.6778 0.0337 asparaginyl-tRNA synthetase<br />
aspC b0928 0.5126 0.0438 aspartate transaminase<br />
aspS b1866 0.5612 0.0181 aspartyl-tRNA synthetase<br />
atpG b3733 1.2411 0.0477 ATP synthase, F1 complex, gamma subunit<br />
b0374 b0374 1.8747 0.0267 putative flagellin structural protein<br />
b1364 b1364 1.9078 0.0056 hypothetical protein<br />
bax b3570 2.7324 0.0137 putative ATP-binding protein<br />
150